|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
EBWS - Bioinformatics
-
RevComOligo
If you want to have your reverse complementary DNA sequence click here…
-
RandomOligoGenerator
If you want to have random DNA, RNA or peptide sequences click here…
-
Motif Searcher
If you want to search for motifs in any DNA, RNA or peptide sequences click here…
-
Restriction Mapping
If you want to search for restriction sites in DNA sequences click here…
-
Prokaryotic ORF Finder
If you want to search for Open Reading Frames (ORFs) in prokaryotic DNA sequences click here…
-
DNA/RNA Mutations
If you want to perform random DNA or RNA mutations at different rates click here…
-
DNA/RNA TM Calculator
If you want to calculate the melting temperature (TM) of DNA or RNA click here…
-
Guide RNA Generator
If you want to generate guide RNA sequences for CRISPR/Cas9 system click here…
-
PCR Annealing Temperature Calculator
If you want to calculate the annealing temperature for PCR reaction click here…
ExBWS-Bioinformatics
-
DNA/RNA Translator
-
AminoCODE
-
Virtual PCR
-
HydropathyPlot-Proteins
If you want to perform a Hydropathy plot of an amino acid sequence click here…
-
Reverse Translator
If you want to perform a Reverse translation of a protein sequence click here…
-
Eukaryotic ORF finder
If you want to search for Open Reading Frames (ORFs) in Eukaryotic DNA sequences click here…
Riboswitch database
-
RSwitch database
To browse the RSwitch database of riboswitches as antibacterial drug targets click here…
Ribozyme generators
-
Oligonucleotide-sensing ribozyme generators
-
Oligonucleotide-sensing ribozyme generator
A generator of slow oligonucleotide-sensing ribozyme with YES Boolean logic function click here…
-
Inverse ribozyme generator
More Applications
-
Bioinformatics Websites
NCBI…; BLAST…; EBI…; PrimerBLAST…; UCSC…; Rfam…; PDB…; KEGG…; BioCYC…;
-
IP Address
If you want to find out your current IP address please click here…
-
Calculator
-
Calendar
An inverse generator of oligonucleotide-sensing ribozyme with YES Boolean logic function
Running the Inverse Folding Generator:
To run the Inverse Folding Ribozyme Generator program
1. Unzip the file you received
2. Open the folder you have extracted the files in the terminal window in Ubuntu
3. Write in the command line: “perl OBSinverse.pl“ and press Enter. The following interface will appear

4. If you press Help the following interface will appear.
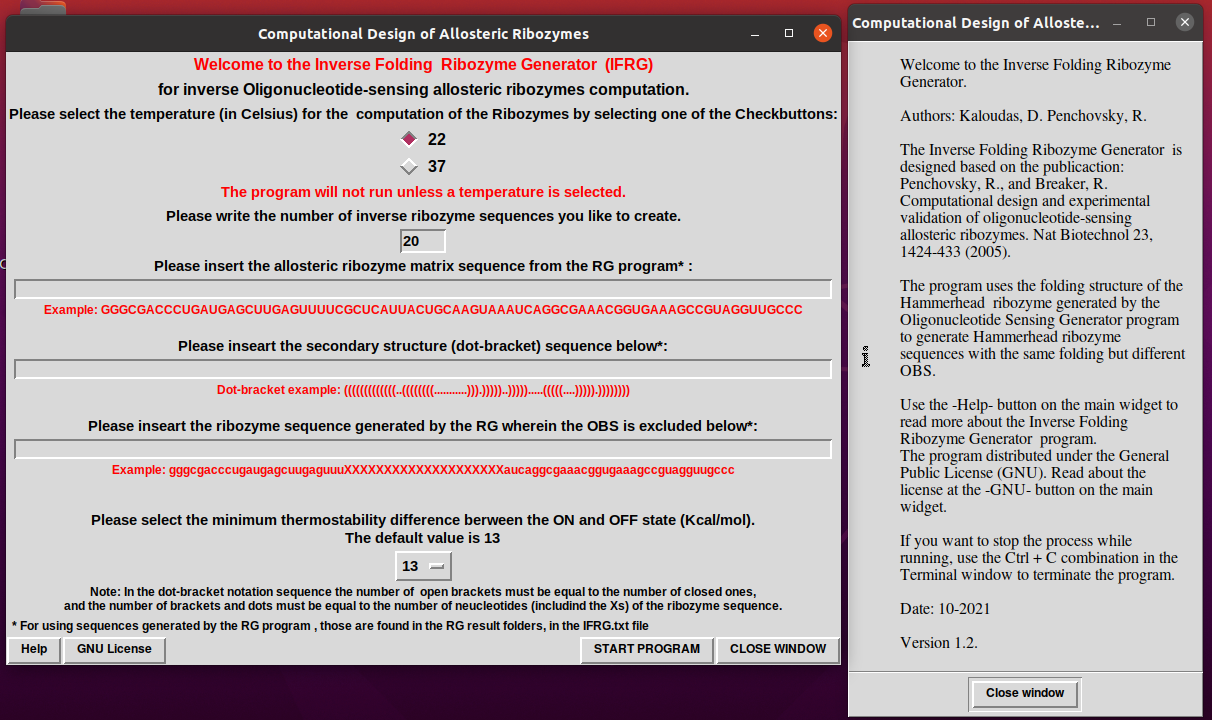
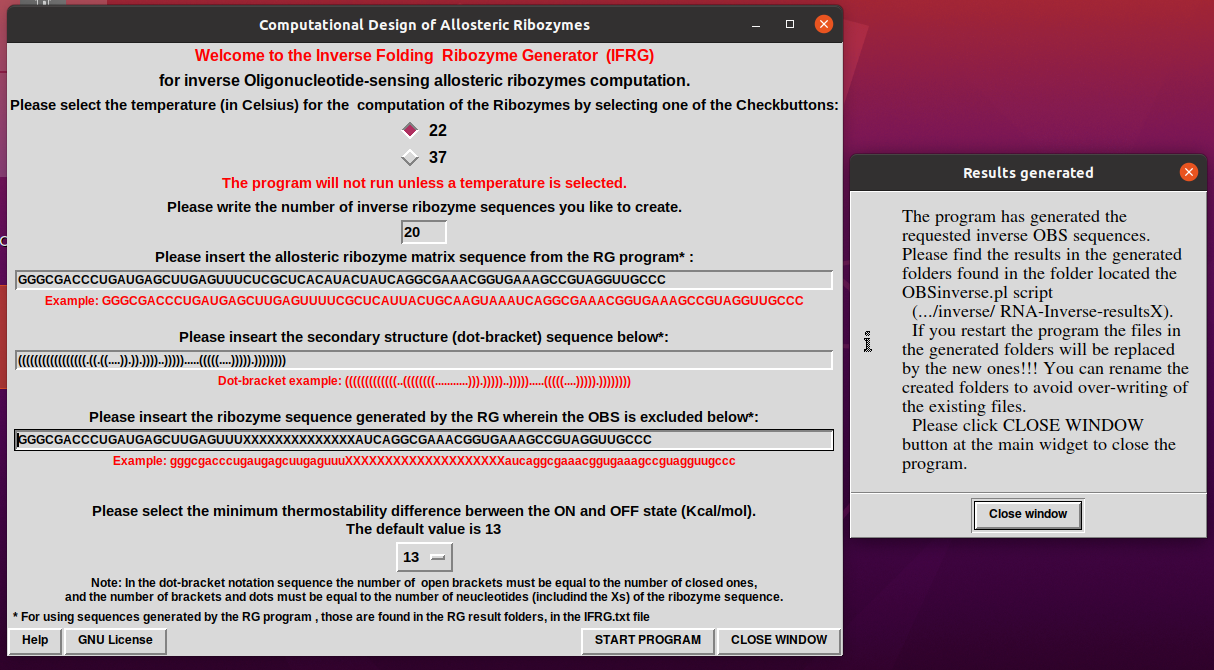
5. When the program finishes with the generation of the sequences a notification widget will pop up.
We have created two computer programs for sequence generation of hammerhead allosteric ribozymes (a oligonucleotide-sensing ribozyme generator, and an inverse ribozyme generator) with YES Boolean logic function that work under Linux/Unix. They are based on algorithms published in Nature Biotechnology and are written in Perl/Tk. The programs are free to use for academic research and you can apply for them by contacting us. The software is under the terms of the GNU General Public License as published by the Free Software Foundation; either version 3 of the License, or at your option any later version. GNU General Public License v3.0 form - GNU Project - Free Software Foundation. For the programs' installation guide click here. That research is funded by grant KP-06-H31/18/13.12.2019 awarded by the Bulgarian National Science Fund (BNSF). This research is published in Computers in Biology and Medicine.