|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Design and Analyses of Non-coding RNAs - Investigating the Role of Non-coding RNAs in Gene Regulation
The discovery of RNA interfering pathways in mammalians including humans raises many hopes for the deployment of novel and efficient tools for gene therapy of viral infections and other wide-spread diseases including cancer by specific targeting of key for the disease` development mRNAs or viral RNAs trough mircoRNAs and RNA silencing pathways. Despite the results achieved in the expression of shRNAs in mammalian cells and the RNA silencing effect demonstrated there are many side effects that spell significant problems facing the application of RNA interfering technology in pharmaceutical industry.
A major problem for the efficient retroviral RNAs silencing such as HIV, is the fact the viral RNAs tend to mutate very fast under the selection pressure of constantly expressed interfering RNA. As a result of that after certain viral generation the specific interfering RNA is not anymore effective towards the virus because of the specific mutations induced by the interfering RNA. In the case of constantly expressed interfering RNA against specific mRNAs strong silencing effect of many non-target mRNAs are observed. To avoid both problems interfering RNAs like shRNAs should be temporary expressed only when needed. This can be achieved by applying RNA-sensing allosteric ribozymes with YES logic function depicted in Fig. 1. An allosteric ribozyme with YES logic function, designed as already described, is constantly expressed into the cell using viral vectors. In the absence of effector RNA the YES gate folds into an inactive state (Fig. 1a). In this conformation the ribozyme`s self-cleavage is inhibited. An effector RNA binds to the ribozyme binding site and turns the ribozyme into an activate (ON) state (Fig. 1b). As a result of the ribozyme`s self-cleavage short-hairpin (sh) RNA is formed (Fig. 1c). The shRNA is converted into double-stranded(ds) RNA with sticky ends by the Dicer enzyme (Fig. 1d). The dsRNA is single- stranded by the RISC complex and used as a template for short interfering RNA to degrade target mRNA or for microRNA to translational repression (Fig. 1e). When the effector RNA, that can be viral and disease indicative RNA, is not present anymore the ribozymes form inactive state again (Fig. 1a) and the shRNA production is held. The restricted time of shRNA production does not allow the targeted viral RNA to mutate under the selection pressure of the shRNA and to escape the silencing effect. The conditional expression of shRNA can reduce the side effects of RNA interfering technology to a minimum and makes it more practically feasible approach to tackle a variety of disorders like viral infection diseases including ADIS and cancer including lymphomas.
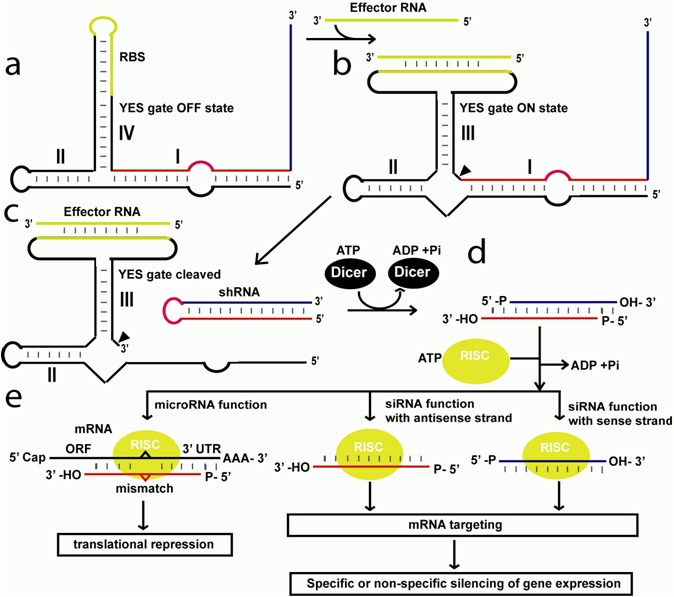
Figure 1. Producing short-hairpin RNAs by RNA-sensing allosteric ribozymes with YES logic function for engineering siRNA and microRNA functions. (a) In the absence of effector RNA the YES gate is designed to fold into an inactive structure where stem IV is formed instead of stem III. (b) The YES switch binds the effector RNA and folds into an active state in which stem III is formed. (c) As a result the ribozyme undergoes self-cleavage and shRNA is produced. (d) Dicer converts shRNA into dsRNA with sticky ends. (e) In the presence of RISC complex the dsRNA is single-stranded and used for gene repression either by RNA silencing via decay of mRNAs or by microRNA function via translational suppression.
References: 1. Robert Penchovsky - Quality Assurance in Healthcare Service Delivery, Nursing and Personalized Medicine: Technologies and Processes. Engineering Gene Control Circuits with Allosteric Ribozymes in Human Cells as a Medicine of the Future – 2012, Publisher IGI Global: DOI: 10.4018/978-1-120-7. 2. Robert Penchovsky - Computational Design of Allosteric Ribozymes as Molecular Biosensors –2014, Biotechnology Advances, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 11,866 3. Nikolet Pavlova, Dimitrios Kaloudas, Robert Penchovsky - Riboswitch distribution, structure, and function in bacteria – 2019, Gene, 0378-1119, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 2,5