|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Design of Programmable Microfluidic Devices for Fully Automated Biochemical Microarrays and Biosensors
Microfluidic technology has been improving rapidly using the expertise accumulated in the etching of tiny patterns during mass production of electronic chips. Many methods based on microfluidics have been already developed in numerous laboratories for various applications including parallel gene synthesis, on-chip PCR, DNA purification, separation, and sequencing, DNA sizing by single molecule detection, light-programmable DNA hybridization chip arrays (Programmieren mit Molekuelen), cellular analysis, and biosensors. Apart from the high-throughput features, microfluidics offers other important advantages such as nanoliter reagent volumes, multi-step processing, on-board mixing of reagents, high reaction rates under flow conditions and possibility for large-scale integration and automation. Despite the significant progress made in recent years in utilizing the potential of microfluidics for biochemical microarrays there was been lacking is a uniform microfluidic platform for fully automated and integrated processing that functions in a parallel fashion.
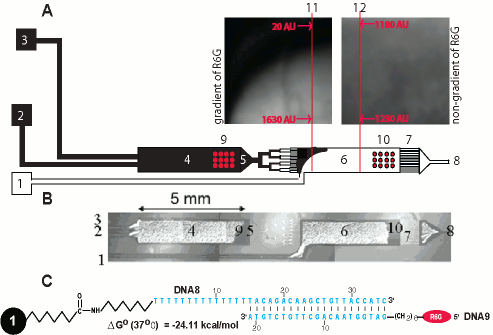
Figure 1. Microchambers connected in a cascade. (A) The microflow reactor consists of three inlet channels (1, 2, and 3), the two chambers (4 and 6) each with its own bead barrier (5 and 7), and one outlet channel (8). Beads are introduced into the first chambers (9) through inlets 1 and 2, and into the second chamber through inlet 3. A Tris buffer solution, pH 8.3, containing rhodomine 6G (R6G) is pumped through inlet 1 at a flow rate of 0.01 ul/min and a R6G free solution is pumped through inlets 2 and 3 at a flow rate of 0.005 ul/min in the microreactor shown below: Fluorescence image from the top part of the second chamber (11). A fluorescence image from the middle part of the second chamber (12) with fluorescence intensity profiles are shown. (B) A white-light image of the microflow reactor used is shown. (C) 35mer DNA8 was immobilized to 15 um beads placed in a microflow reactor. Two different 5?-end R6G labeled are used. DNA9 is a perfectly matching oligomer.
Herein, the general design of microfluidic for multi-step sequence-specific nucleic acid selection is built on microfluidic platform in cascading microchambers that is fully programmable and automated. The selection scheme can be designed to work in parallel, taking into account previous work on developing microfluidic devices for an isothermal DNA selection.
The mixing efficiency in two cascading microreactor (Fig. 1) is shown. To estimate the mixing efficiency in the second chamber NaOH solution was delivered through inlets 2 and 3. A 500 mM Tris-acetate buffering solution, containing R6G was delivered simultaneously through inlet number 1 (Fig. 1A). Fluorescence images from the top part of the second chamber and from its middle part were obtained. There is a clear border between the two pumped solutions in the top part of the second chamber (Fig. 1A). The border, between the two fluids disappears in the middle part of the chamber pursuant to an almost complete mixing between the two solutions under the applied flow rate (Fig. 1A). A white light image of the cascading microchambers is shown in Fig. 1B.
Video 1. Real time video of DNA/DNA hybridization kinetics and denaturation by a NaOH solution on beads under flow conditions. The details of this experiment are submitted for a publiction.
A novel microfluidic architecture is described herein for fully automated and integrated biopolymer arrays. The design is based on the pH-reversible chemistry for an isothermal DNA hybridization transfer between cascade microchambers and the programmable synthetic DNA immobilization. The established inhibition of the immobilization reaction guarantees high-specificity in DNA attachment. Both processes, the immobilization and the selection, are fully integrated and automated, and take place in parallel. A microfluidic architecture of the microfluidic architecture described contains 2 supply horizontal channels, 12 horizontal channels, each with 8 cascade microchambers, crossed by 7 vertical supply channels at 84 crossing points (Fig. 2). There is a mixing structure after each crossing point that combines vertical and chaotic mixers followed by a chamber, being a part of the horizontal channels. The number of chambers is 96. Each chamber has a bead barrier at the bottom. Paramagnetic beads can be delivered to each chamber via the vertical channels except for the chambers of the first row, the beads being delivered by the horizontal channels. There is one valve at both ends of each vertical supply cannel as a total number of 14. Note that the number of valves, inlets and outlets increases only linearly while the number of chambers increases quadratically. That makes the described design more scalable, since large numbers of chambers are much easier to integrate compared to either valves or inlets and outlets. The microfluidic architecture can be used for creating DNAs, RNAs, polypeptides and cellular microfluidic arrays. The approach presented is universal as every type of biopolymer can be selectively immobilised to the beads placed in different chambers or to their surfaces by use of almost any cross-linking chemistry including photo-dependent DNA immobilization in microfluidics. That makes the biochip`s production chemistry-independent and very suitable for HTS arrays for drug discovery.
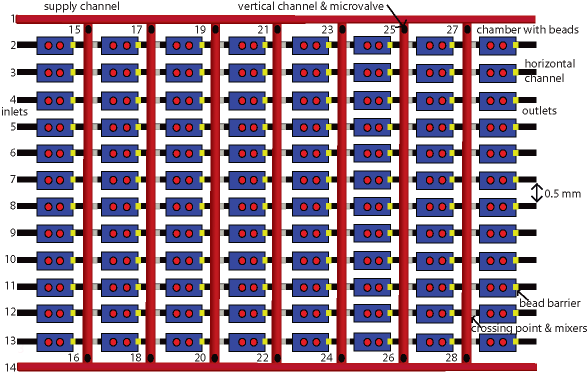
Figure 2. A microfluidic architecture for programmable and parallel biopolymer immobilization and selection. Microfluidic architecture. There are 2 supply channels (1 & 14), 12 horizontal channels (2-13) crossed by seven vertical channels (84 crossing points secured by small connecting channels) with mixers and 14 microvalves (15-28). After each crossing point, there is a mixing structure. There are 96 chambers with an etched bead barrier at the bottom of each one. Carboxyl-coated magnetic beads are to be incorporated in all 96 chambers.
References: 1. Robert Penchovsky, E. Birch-Hirschfeld and John S. McCaskill - End-specific covalent photo-dependent immobilisation of synthetic DNA to paramagnetic beads – 2000, Nucleic Acids Res., 1362-4962, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 9,338 2. John S. McCaskill, Robert Penchovsky, Marlies Gohlke, Jorg Ackerman and Thomas Rücker - Steady flow micro-reactor module for pipelined DNA computations – 2001, Lecture Notes in Computer Sciences, 03029743, Q4 – 12 т. (Computer Science), IF – 1,2 3. Robert Penchovsky and John S. McCaskill - Cascadable hybridisation transfer of specific DNA between microreactor selection modules – 2002, Lecture Notes in Computer Sciences, 03029743, Q4 (Computer Science), IF – 1,2 4. Robert Penchovsky - Programmable and automated bead-based microfluidics for versatile DNA microarrays under isothermal conditions – 2013, Lab on a chip, 14730197, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 6,41 5. Robert Penchovsky - Automated DNA hybridization transfer with movable super-paramagnetic microbeads in a microflow reactor – 2019, Biosensors and Bioelectronics, 0956-5663, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 8,17 6. Robert Penchovsky - An Integrated DNA Selection in Micro-flow Reactors as an Approach for Molecular Computation and Diagnostics. Ph.D. thesis, Universität zu Köln -2003. 7. Robert Penchovsky, Relle, M., Rücker, T, McCaskill, J.S - Programmieren mit Molekülen – 1999, Der GMD-Spiegel