|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Computational Design of Allosteric Ribozymes that function as Boolean logic gates in vitro and in vivo
Allosteric RNAs operate as molecular switches that alter folding and function in response to ligand binding. Common types of natural allosteric RNAs are riboswitches, and designer RNAs with similar properties can be created by RNA engineering. We describe a computational approach for designing allosteric ribozymes triggered by binding oligonucleotides. Four universal types of RNA switches possessing AND, OR, YES, and NOT Boolean logic functions were created in a modular form, which allows ligand specificity to be changed without altering the catalytic core of the ribozyme. All computationally designed allosteric ribozymes were synthesized and experimentally tested in vitro. Engineered ribozymes exhibit >1,000-fold activation, demonstrate precise ligand specificity, and can function in molecular circuits wherein the self-cleavage product of one RNA triggers the action of a second. This engineering approach provides a rapid and inexpensive way to create allosteric RNAs for constructing complex molecular circuits, nucleic acid computing systems, and gene control elements. The approach is also a subject of a pending patent application. In addition, this work has been acknowledged by News and Views publications in Nature Biotechnology and Nature Methods.
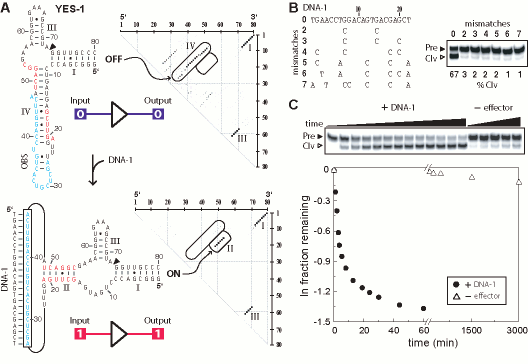
Figure 1. Design and characterization of an oligonucleotide-specific RNA switch possessing YES logic function. (A) Secondary structure models for the most stable conformers as computed using the partition function algorithm in the absence (OFF) or presence (ON) of a 22-nucleotide DNA effector. The effector-binding site (light blue) is joined to nucleotides 10.1 and 11.1 of the hammerhead core via eight- and six-nucleotide linkers. In the ON state, most of these linker nucleotides are predicted to form an extended stem II structure (green). To the right of each model is a dot matrix plot wherein larger points reflect a greater probability of base pairing. Encircled points reflect the main differences in predicted structures between the OFF (stem IV) and ON (stem II) states. Nucleotides 1 through 79 are numbered from 5` to 3` across the top and right of the plots. Schematic representations of the logic states of the constructs are shown in this and subsequent figures. (B) Selective activation of ribozyme self-cleavage by an effector DNA complementary to the oligonucleotide binding site (OBS). Radiolabeled ribozymes (5` 32P, Pre) undergo self-cleavage only with the perfectly matched DNA effector (0 mismatches) and the resulting radiolabeled cleavage fragment (Clv) is separated from the precursor by denaturing 10% PAGE. Products were visualized and cleavage yields were quantitated by PhosphorImager. Reactions were conducted for 10 min at 23oC in standard buffer conditions (100 mM Tris-HCl [pH 8.0 at 23oC] and 20 mM MgCl2). (C) Kinetics of ribozyme (1 uM) self-cleavage in the presence (+) of perfectly matched 22-nt effector DNA (3 uM) and in the absence (-) of effector DNA. Gel image is as described in B. Plot using data derived from the gel depicts the natural logarithm of the fraction of RNA remaining uncleaved versus time.
Molecules with attributes of YES logic function must remain inactive unless receiving a single molecular impulse that triggers activity. One of these constructs, termed YES-1, is predicted to form the desired OFF- and ON-state structures in the absence and presence, respectively, of a 22-nt effector DNA (DNA 1; Fig. 1A). In its inactive conformation, the nucleotides within the OBS are proposed to form a 'stem IV' structure. Stem IV involves extensive base-pairing interactions with portions of the hammerhead core and with most nucleotides that would otherwise form the stem II structure required for ribozyme activation. In the presence of DNA-1, a major portion of the nucleotides in stem IV would become sequestered by intermolecular base pairing, and nucleotides that can participate in stem II formation are liberated. The results of the computational assessment of the structure-forming potential of this construct are visually represented by dot matrix plots (Fig. 1A, right).It is apparent from these plots that the probability of forming stem IV or several base-pairing alternatives is high, whereas there is no indication that stem II has any reasonable chance of forming. In contrast, repeating the computation in the presence of the DNA effector drastically reduces the probability that stem IV can form, and thus increases the probability that stem II will be formed. It is notable that the probabilities for forming hammerhead stems I and III remain largely unaffected by the presence or absence of the effector DNA. In a subsequent stage of the design procedure, the secondary structure adopted for the YES-1 ribozyme was used as a basis to compute the folding properties of additional allosteric ribozyme candidates that are likely to have very similar RNA energy folding landscapes but that carry different OBS sequences that respond to different effector DNAs
To estimate the dynamic range for allosteric activation, or the total range of rate constant enhancement brought about by effector binding, a time course for ribozyme self-cleavage was conducted in the presence and absence of the matched effector DNA (Fig. 1C). In the presence of the effector, the apparent rate constant observed for ribozyme activity (apparent kobs) is ~1.1 x 10-1 min-1, whereas the apparent kobs for the ribozyme in the absence of effector DNA is ~1.6 x 10-5 min-1. These results indicate that the allosteric dynamic range is nearly 7,000 fold, and the maximum rate constant is within 10-fold of the typical maximum activity for the unmodified hammerhead ribozyme core (~1 min-1) measured under similar conditions. Furthermore, the stability of the OFF-state structure is not so extreme or so rapidly adopted that activation by the effector DNA is precluded when a reaction buffer (Fig. 1B) and DNA are introduced simultaneously to the YES-1 ribozyme.
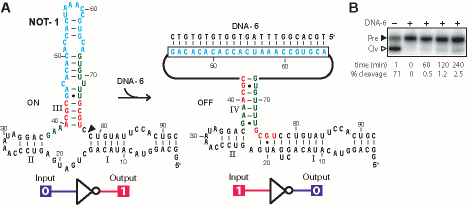
Figure 2. Design and characterization of NOT-1 based on an extended hammerhead ribozyme. (A) Secondary structure models for the most stable conformers predicted in the absence (ON) and presence (OFF) of a 23-nucleotide effector. (B) Deactivation of NOT-1 by a DNA complementary to the OBS. Ribozyme assays were conducted at 37oC in a solution containing 2 mM MgCl2, 100 mM KCl, 25 mM NaCl, 50 mM Tris-HCl (pH 7.5 at 37oC). Ribozyme reactions were initiated by the addition of MgCl2 after pre-incubating NOT-1 and 10-fold excess DNA-6 (when present) for 5 min in Mg2+-free reaction buffer.
An extended natural hammerhead ribozyme from Schistosoma mansoni, was used as the parent construct for the design of a ribozyme that is deactivated by allosteric interactions with oligonucleotides. This ribozyme exhibits faster RNA cleavage kinetics and requires lower concentrations of Mg2+ to trigger activity. Allosteric constructs derived from parental ribozymes with these properties are more likely to function in vivo where divalent ion concentrations are low and where fast ribozymes might be needed. Extended hammerhead ribozymes exhibit improved function because they form a tertiary structure between the loop sequences of stem II and a bulge within stem I. Therefore, the OBS was relocated to stem III for the design of a construct that functions as a NOT gate so this critical ribozyme tertiary-structure contacts would not be disrupted. The resulting design, termed NOT-1 (Fig. 2A) is predicted to form a single major ON state structure (Ep = -30.55 kcal mol-1) in the absence of effector DNA-6 (23 nt), and is predicted to form a single major structure in its effector-bound OFF state that has an extended stem I and a disrupted stem III (Ep = -25.46 kcal mol-1). If the NOT-1 construct functions as predicted and self-cleaves in the absence of effector DNA, then the preparation of the RNA is expected to pose a problem because the ribozyme could undergo self-cleavage during transcription in vitro. To avoid this outcome, DNA templates corresponding to the NOT-1 RNA were transcribed in the presence of 10 uM DNA-6 and 10 uM of the antisense oligonucleotide CTCATCAGC. The latter DNA is complementary to nucleotides 15 through 23 of the NOT-1 hammerhead core. Although the NOT-1 RNA exhibited only ~25% self-cleavage when produced by transcription under these conditions (data not shown), the RNA exhibits robust self-cleavage activity (kobs >1 min-1) when incubated in the absence of DNA-6 (Fig. 2B). Similarly, addition of excess effector oligonucleotide to a NOT-1 ribozyme assay causes strong inhibition.
References:
1. Robert Penchovsky & Ronald R Breaker - Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes – 2005, Nature Biotechnology, 10870156, Q1(Biochemistry, Genetics and Molecular Biology), IF – 43,5
2. Robert Penchovsky - Engineering integrated digital circuits with allosteric ribozymes for scaling up molecular computation and diagnostics – 2012, ACS Synth Biol, 21615063, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 5,382
3. Robert Penchovsky, Kostova GT. - Computational selection and experimental validation of allosteric ribozymes that sense a specific sequence of human telomerase reverse transcriptase mRNAs as universal anticancer therapy agents – 2013, Nucleic Acid Therapeutics, Q1 (Biochemistry, Genetics and Molecular Biology) IF – 2,338
4. Robert Penchovsky - Computational design and biosensor applications of small molecule-sensing allosteric ribozymes – 2013, Biomacromolecules, 1525-7797, Q1 (Chemical Engineering), IF – 5,77
5. Robert Penchovsky - Computational Design of Allosteric Ribozymes as Molecular Biosensors – 2014, Biotechnology Advances, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 11,866
6. Robert Penchovsky - Quality Assurance in Healthcare Service Delivery, Nursing and Personalized Medicine: Technologies and Processes. Engineering Gene Control Circuits with Allosteric Ribozymes in Human Cells as a Medicine of the Future – 2012, Publisher IGI Global: DOI: 10.4018/978-1-120-7.
7. Ronald R. Breaker & Robert Penchovsky - US20110288826 Computational design of ribozymes - 2011, WIPO
8. Robert Penchovsky & Martina Traykovska - Synthetic Approaches to Biology: engineering gene control circuits, synthesizing, and editing genomes, Emerging Research on Bioinspired Materials Engineering (book chapter) – 2016, IGI Global, 9781466698116
9. Robert Penchovsky - Handbook of Research on Nanoscience, Nanotechnology, and Advanced Materials (book chapter) – 2014, Engineering science reference: An Imprint of IGI Global, 146665824X
10. Adam A Margolin & Milan N Stojanovic - Boolean calculations made easy (for ribozymes) - Nature Biotechnology - 2005, IF:43.113
11. Michael Eisenstein - Computational evoluation offers riboswitch solution - Nature Methods - 2005, IF:25.56
12. Clare Sansom - When is an enzyme not a protein? - RSC Chemistry World - 2016
13. Dimitrios Kaloudas & Robert Penchovsky - An allosteric ribozyme generator and an inverse folding ribozyme generator: Two computer programs for automated computational design of oligonucleotide-sensing allosteric hammerhead ribozymes with YES Boolean logic function based on experimentally validated algorithms - Computers in Biology and Medicine - 2022, IF - 4.5, Q1, Scopus
14. Robert Penchovsky, Georgi Miloshev, Nilolet Pavlova, Katya B. Popova, Lozena A. Otcheva and Martina Traykovska, Book chapter 8: Small RNA-based systems for sensing and therapeutic applications - New Frontiers and Applications of Synthetic Biology, Elsevier - 2022