|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
High-throughput Screening Assays for Antibacterial Drug Discovery Employing RNA Targets
Bacterial riboswitches are natural aptamers in UTR region of a mRNA molecule that can directly bind a small target molecule and effect the gene expression. For time being 12 bacterial riboswitch classes are discovered that regulate gene expression through translation prevention, transcription termination, and self-cleavage. The encounter of one or more riboswitch classes in 15 Human bacterial pathogens raises the hopeful of using them as novel targets for antibacterial drug discovery. The demand for novel antibiotics is constantly increasing since more Human bacterial pathogen stems are becoming resistant to long time used antibiotics. In this regard novel antibacterial drug targets may be valuable to pharmaceutical industry and may be a commercial success.
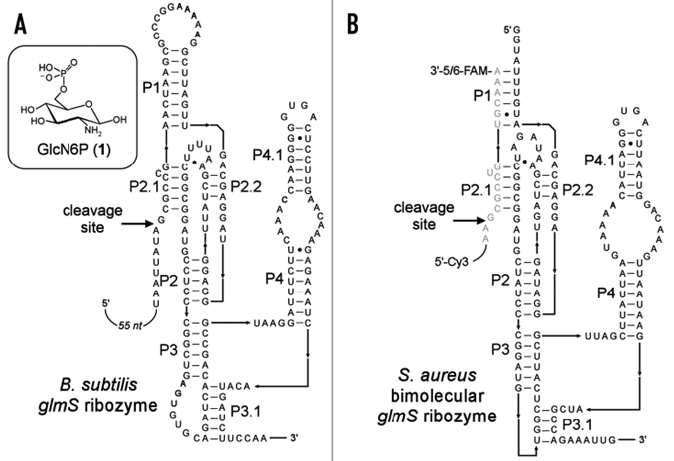
Figure 1. The secondary structures of wild-type and FRET glmS ribozymes. (A) An unimolecular glmS ribozyme from B subtilis. Arrow identifies the site of ribozyme-mediate cleavage stimulated by GlcN6P (1). (B) A bimolecular glmS ribozyme construct derived from S. aureus. This construct differs from the wild-type glmS ribozyme due to truncation of the P1 stem and the use of a 15-nucleotide substrate RNA (shown in grey). The substrate is labeled with a Cy3 acceptor at its 5`-end and a 5/6-FAM donor at its 3`-end.The FRET ribozyme construct was used for HTS arrays.
I have been involved in the development and application of an assay for glmS riboswitch. The glmS riboswitch is a ribozyme that cleaves itself in the presence of glucose amine 6-phosphate. It is present in 4 Human bacterial pathogens. It was straight forward to develop a cleave-based high-throughput screening assay for the glmS riboswitch using FRET (Fig. 1). The rest riboswitch classes only bind a small target molecule without self-cleavage. Thus, the development of a cheap and effective biochemical high-throughput screening assay for 11 riboswitch classes is not a straight forward task. The application of computational high-throughput screen assays for those riboswitch classes may be a valuable option for the riboswitch-based antibacterial drug discovery. The automated pipeting system used for HTS arrays is shown in Fig.2.

Figure 2. A pipeting robot system for fully automated high-throughput screening arrays.
References: 1. Kennet Blount, Izabela Puskarz, Robert Penchovsky, Ronald Breaker - Development and application of a high-throughput assay for glmS Riboswitch Activators – 2006, RNA Biolоgy, 15558584, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 5,546 2. Robert Penchovsky, Stoilova C.C. - Riboswitch-based antibacterial drug discovery using high-throughput screening methods – 2013, Expert Opinion on Drug Discovery, 1746-0441, Q1 (Pharmacology, Toxicology and Pharmaceutics) , IF – 4,676 3. Robert Penchovsky & Martina Traykovska - Designing drugs that overcome antibacterial resistance: where do we stand and what should we do? – 2015, Expert opinion on drug discovery, Q1 (Pharmacology, Toxicology and Pharmaceutics), IF – 4,66 4. Aikaterini Valsamatzi-PanagiotouKatya B. Popova & Robert Penchovsky - Chapter 9: Drug Discovery for Targeting Drug Resistant Bacteria – 2020, Sustainable Agriculture Reviews 46, Mitigation of Antimicrobial Resistance Vol. 1, Tools and Targets:. 5. Aikaterini Valsamatzi-PanagiotouKatya B. Popova & Robert Penchovsky - Chapter 1: Strategies for prevention and containment of antimicrobial resistance – 2020, Sustainable Agriculture Reviews 49 Mitigation of Antimicrobial Resistance Vol. 2, Natural and Synthetic Approaches.