|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Molecular Computing - Employing Nucleic Acids as Molecular Computing Devices
With a large quantity of DNA, bio-molecular-based computers may offer the possibility that massive parallelism could be used for solving NP complete problems in polynomial time. Instances of NP complete problems, such as the maximum clique problem and the SAT problem have been solved using DNA/DNA hybridization. A key question in DNA computing is the fidelity of the basic operations employed and the scalability of the computation as a whole. When DNA/DNA hybridization is used as a basic computational operation, the accuracy of the computation will depend on the ability to discriminate between perfectly matching hybrids (the bits of the library and their complementary oligomers) and those with mismatches. In this regard, the quality of the DNA code design is playing a critical role in the fidelity of the computation. The problem of designing sets of modular RNA and DNA sequences, which hybridize in a predefined way, is fundamental not only for molecular computing but also for many applications in contemporary molecular biology and nanotechnology such as strand selection, PCR and isothermal amplification reactions, DNA-chip arrays, ribozyme design, and self-assembly of DNA. We have designed a 12-bit DNA library that encodes binary information by our own software based on thermodynamics constrains. The results show a high level of specific hybridization achieved for all library words under identical conditions (Table 1). The approach can be used for designing molecular logic gates for biosensor arrays that function in parallel.
Table 1. A 12-bit DNA library that encodes binary information.The melting points for all words are calculated thermodynamically for 50 mM NaCl and 1 uM oligomer using two different thermodynamic data. The highest and the lowest melting points are 2.9+/- oC (without taking into account the GC and AT init. w/term.) according to Breslauer et al. (1986) (Melting points-1) and Allawi and Santa Lucia (1997) (Melting points-2).
| N | Bits | 5'- 3' Word sequences |
5'- 3' Capture sequences |
Melting points-1 | Melting points-2 |
|---|---|---|---|---|---|
| 1 | 1.1 | CCATCACTACCTTCAT | ATGAAGGTAGTGATGG | 45.3 | 46.8 |
| 2 | 1.0 | TCCTCTATCATCCTCA | TGAGGATGATAGAGGA | 46.6 | 46.5 |
| 3 | 2.1 | TCCCTATTCACTCTCT | AGAGAGTGAATAGGGA | 44.6 | 46.7 |
| 4 | 2.0 | CACACCTCAACTTCTT | AAGAAGTTGAGGTGTG | 44.9 | 48.0 |
| 5 | 3.1 | ACTTCCCTTCTACACA | TGTGTAGAAGGGAAGT | 44.8 | 48.1 |
| 6 | 3.0 | CACCATCCTTATCTCA | TGAGATAAGGATGGTG | 46.7 | 46.4 |
| 7 | 4.1 | TCTCTCAATCCACTTC | GAAGTGGATTGAGAGA | 45.6 | 46.6 |
| 8 | 4.0 | TACAATCCCACACTTT | AAAGTGTGGGATTGTA | 45.6 | 46.6 |
| 9 | 5.1 | TCTCTTCCTCTTACCA | TGGTAAGAGGAAGAGA | 45.8 | 47.0 |
| 10 | 5.0 | TCATACCTAACTCCCT | AGGGAGTTAGGTATGA | 44.6 | 46.7 |
| 11 | 6.1 | CTCATCTTAACCACCT | AGGTGGTTAAGATGAG | 44.7 | 46.7 |
| 12 | 6.0 | ACCATTACTTCAACCA | TGGTTGAAGTAATGGT | 45.6 | 46.6 |
| 13 | 7.1 | TTCTACAACCTACCCT | AGGGTAGGTTGTAGAA | 44.4 | 47.3 |
| 14 | 7.0 | TCCAACTTAACACTCC | GGAGTGTTAAGTTGGA | 45.3 | 47.3 |
| 15 | 8.1 | ACCTTTACCCTATCCT | AGGATAGGGTAAAGGT | 46.2 | 47.1 |
| 16 | 8.0 | ACACCCTAACAATCAA | TTGATTGTTAGGGTGT | 45.6 | 46.6 |
| 17 | 9.1 | CACCCATTCCTAATAC | GTATTAGGAATGGGTG | 45.4 | 45.2 |
| 18 | 9.0 | TCCTACACAAACATCA | TGATGTTTGTGTAGGA | 43.8 | 46.3 |
| 19 | 10.1 | ATTCTCACTCACAACC | GGTTGTGAGTGAGAAT | 44.6 | 47.8 |
| 20 | 10.0 | ACCACTCCAATAACTC | GAGTTATTGGAGTGGT | 44.2 | 47.0 |
| 21 | 11.1 | TCCTACTCTCCAATCA | TGATTGGAGAGTAGGA | 46.4 | 47.1 |
| 22 | 11.0 | TCTTTCACACATCCAT | ATGGATGTGTGAAAGA | 46.3 | 46.7 |
| 23 | 12.1 | ACACCATTTCACCTAA | TTAGGTGAAATGGTGT | 45.6 | 46.6 |
| 24 | 12.0 | ACACTAATCCTCCAAC | GTTGGAGGATTAGTGT | 44.2 | 47.0 |
Molecules with attributes of AND logic function must remain inactive unless receiving two separate molecular impulses that trigger activity. Candidate RNA constructs possessing AND logic function triggered by 16-nt effector DNAs were designed using the same principles and computational procedures used to identify candidate YES RNA switches. However, additional steps were added to permit computation of four different structural states with high stability. As with the YES gate computations, one of the structural states must permit the formation of the active hammerhead core, in this case, only when presented with two effector DNA sequences. The remaining three states should not permit ribozyme function, even if either of the two effector DNAs is present independently. Our computational search efforts indicate that many thousands of ribozymes with the same AND gate properties can be generated. Many molecular logic gates with various functions can be designed to work in parallel.
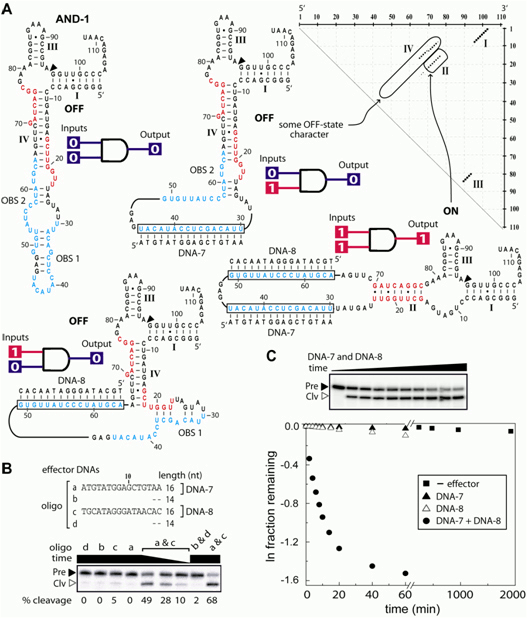
Figure 1. Design and characterization of AND-1, an oligonucleotide-specific molecular switch that possesses AND logic function. (a) AND-1 is designed to form the active hammerhead structure and self-cleave only when presented simultaneously with its two corresponding effector DNAs (DNA-7 and DNA-8). The dot matrix plots for the ON state showing some character of the OFF states (stem IV) is depicted. (b) Activation of AND-1 self-cleavage requires both full-length DNA-7 and DNA-8 effectors. Maximum incubation time is 60 min. (c) Kinetics of AND-1 self-cleavage under various combinations of effector DNAs.
References: 1. Robert Penchovsky and Jorg Ackerman - DNA library design for molecular computation – 2003, J Comput Biol., 10665277, Q2 – 20 т. (Biochemistry, Genetics and Molecular Biology), IF – 1,89 2. Robert Penchovsky & Ronald R Breaker - Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes – 2005, Nature Biotechnology, 10870156, Q1(Biochemistry, Genetics and Molecular Biology), IF – 43,5 3. Robert Penchovsky - Engineering integrated digital circuits with allosteric ribozymes for scaling up molecular computation and diagnostics – 2012, ACS Synth Biol, 21615063, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 5,382 4. Robert Penchovsky - An Integrated DNA Selection in Micro-flow Reactors as an Approach for Molecular Computation and Diagnostics. Ph.D. thesis, Universität zu Köln -2003.