|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Engineering Gene Regulatory Networks for Exogenous Control of Gene Expression and Gene Therapy
An application of the allosteric ribozymes as Boolean logic gates in vivo can open up new avenues to the development of universal strategies for exogenous control of gene expression and gene therapy of cancer. The approach is also a subject of a pending patent application and can be used as a universal tool for reprogramming the cell fate by inducing apoptosis or cell differentiation with conditional exogenous expression of key proteins like p53. This may have many far-reaching applications to the pharmaceutical and biotech industries.
A designer construct that contains two allosteric ribozymes and a p53 gene, can be delivered by viral expression vector into a mix population of cancerous and normal cells. The construct expresses a mRNA that encodes exogenous p53 and two allosteric ribozymes with a NOT logic function. Both ribozymes are designed to sense the presence of two indicative of the cancer mRNAs (Fig. 1). The pathological phenotype of the cell is defined by the co-expression of the two cancer indicative RNAs. The following general anticancer gene therapy shown in Fig. 1 can be employed to induce apoptosis in all cancer cells when both cancer indicative RNAs are presented (Fig. 1b), and to spare the normal cells when either or both cancer indicative RNAs are not presented (Fig. 1a). The proposed strategy for exogenous control of gene expression is general and can be applied in many other regulatory circuits in vivo.
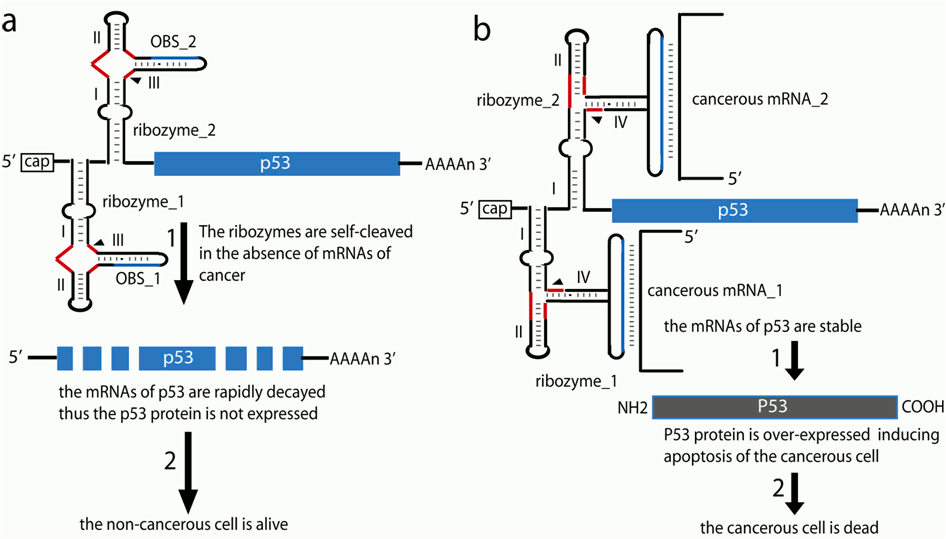
Figure 1. Re-programming the cell fate applying allosteric ribozymes with NOT Boolean logic function as a universal anticancer gene therapy tool.(a) The cell is normal. In the absence of one of the two or both cancerous mRNAs, the p53 mRNA is decapitated due to the function of the ribozymes (1). As a result the mRNA of p53 is rapidly decayed and the exogenous p53 protein is not expressed. The normal cell is alive. (b) The cell is cancerous and both cancer indicative mRNAs are presented. They bind to the corresponding sites of the ribozymes and suppress their function (cleavage). As a result the p53 mRNA is not rapidly decayed (1) and the exogenous p53 protein is over-expressed, which induces apoptosis of the cancerous cell (2). The effector RNAs can be identical to both ribozymes.
One promising real-world application of this approach would be addressing some forms of liver cancer and MCL because we can transform liver and B-cells in vivo with high efficiency maintaining constant viral expression for at least two weeks. In addition, there are cancer indicative mRNAs that are over expressed during the early stage of liver cancer and MCL development. The p53 is not the only protein that must be over-expressed always to induce apoptosis. For instance, it is observed selection against PUMA gene expression in Myc driven B-cell lymphomagenesis, which means that exogenous overexpression of PUMA in such cells may be more suitable than that of p53. Moreover, selective over-expression of exogenous Arf in some lymphoma cells can lead to suppression of Myc-driven lymphomagenesis.
Boolean logic gates with a YES function can be used to turn exogenous gene expression OFF as shown in Fig. 2. In this case the expression of mRNAs, that are indicative of the wild-type (wt) state of the cell, induce the self-cleavage of the ribozyme and prevent the over-expressed of exogenous p53 (Fig. 2a) and guaranties the survive of wt-cell. Otherwise, without one or both wt-indicative RNAs, the cell is malignant and will over-expressed the p53 inducing apoptosis(Fig. 2b). For instance, it is known that Atm deficiency can affect both apoptosis and proliferation to augment Myc-induced lymphomagenesis.
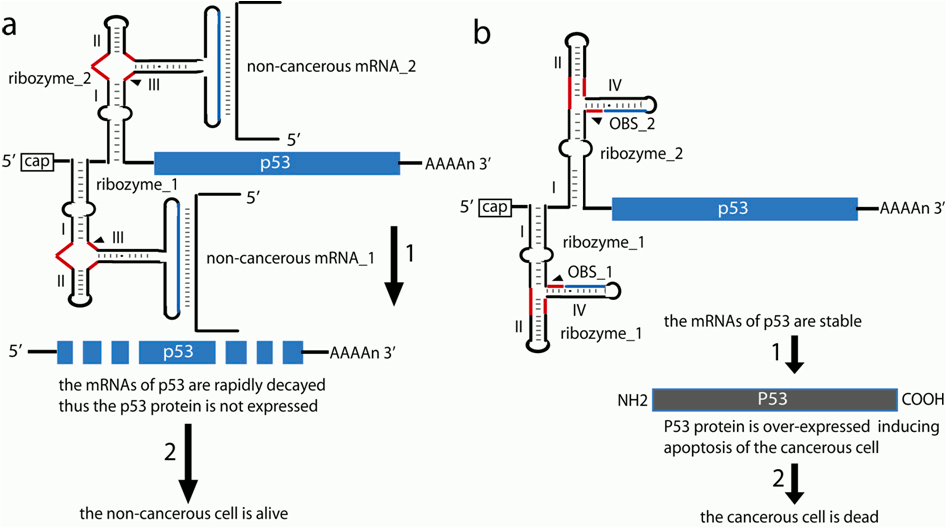
Figure 2. Re-programming the cell fate using YES allosteric ribozymes as a universal anticancer gene therapy tool. (a) The cell is non-cancerous because the presence of two indicative mRNAs. In the presence of both mRNAs, the p53 mRNAs are decapitated due to the function of the two ribozymes (1). As a result the mRNAs of p53 are rapidly depredated and the exogenous P53 protein is not an expression. The normal cell is alive. (b) The cell is cancerous and both indicative mRNAs are not presented. The ribozymes with YES Boolean logic function are inactive. As a result the p53 mRNA is not rapidly decayed (1) and the exogenous P53 protein is over-expressed, which induce apoptosis and the cancer cell dies (2).

References:
1. Robert Penchovsky & Ronald R. Breaker - Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes – 2005, Nature Biotechnology, 10870156, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 43,5.
2. Robert Penchovsky - Quality Assurance in Healthcare Service Delivery, Nursing and Personalized Medicine: Technologies and Processes. Engineering Gene Control Circuits with Allosteric Ribozymes in Human Cells as a Medicine of the Future – 2012, Publisher IGI Global: DOI: 10.4018/978-1-120-7.
3. Robert Penchovsky & Kostova G.T. - Computational selection and experimental validation of allosteric ribozymes that sense a specific sequence of human telomerase reverse transcriptase mRNAs as universal anticancer therapy agents – 2013, Nucleic Acid Therapeutics, Q1 (Biochemistry, Genetics and Molecular Biology) IF – 2,338
4. Robert Penchovsky - Present and Future RNA-based Approaches to Medical Genomics – 2013, Journal of Clinical & Medical Genomics, 2472-128X, IF – 0,58
5. Robert Penchovsky & Martina Traykovska - Synthetic Approaches to Biology: engineering gene control circuits, synthesizing, and editing genomes, Emerging Research on Bioinspired Materials Engineering (book chapter) – 2016, IGI Global, 9781466698116
6. Robert Penchovsky - Computational design and biosensor applications of small molecule-sensing allosteric ribozymes – 2013, Biomacromolecules, 1525-7797, Q1 (Chemical Engineering), IF – 5,77
7. Robert Penchovsky - Computational Design of Allosteric Ribozymes as Molecular Biosensors –2014, Biotechnology Advances, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 11,866
8. Georgi Miloshev, Martina Traykovska, Dimitrios Kaloudas and Robert Penchovsky - Engineering a plasmid as a reporter system for quantifying gene expression in Escherichia coli, Comptes rendus de l'Académie bulgare des Sciences -2022…IF:0.38, (Q2, Scopus). The sequence of our plasmid pRS414ge is given here.