|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
Robert's Research
-
Computational Design of Allosteric Ribozymes
-
Engineering Gene Regulatory Networks
-
Design and Analyses of Non-coding RNAs
investigating the role of non-coding RNAs in gene regulation…
-
Molecular Computing
-
Ribozyme-based Molecular Circuit
-
Computational Drug Design
applying virtual high- throughput screening assays and rational design…
-
High-throughput Screening Assays
-
Targeting specific RNAs by antisense oligonucleotides
Inhibition of bacterail growth by targeting specific RNAs by antisense oligo- nucleotides (ASOs)…
-
Design of Programmable Microfluidic Devices
-
Software Design for Bionformatics
Lab's members
-
Professor Robert Penchovsky, Ph.D.
-
Assistant Professor Martina Traykovska, Ph.D.
-
Assistant Professor Nikolet Pavlova, Ph.D.
-
researcher Dimitrious Kaloudas, Ph.D.
-
Antoniya Georgieva, M.Sc.
-
Vanya Dyakova, M.Sc.
Main Grant Awards
-
grant:DDVU02/5/2010
Design and applications of RNA biosensors in vitro and in vivo…
-
grant:DN13/14/20.12.2017
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
grant:KP-06-H31/18/13.12.2019
-
grant:KP-06-H63/1/13.12.2022
-
grant:4011/05.07.2023
-
grant:70-123-194/12.02.2024
Creation of software systems for computer-aided design of rapid allosteric ribozymes that sense the presence of sequence-defined oligonucleotides and a database of clinically relevant human genetic variation (budget: 102300 EURO )...
Research Awards
-
Dr. Penchovsky's outstanding scientist award, 2023
7th Edition of International Research Awards on SENSING TECHNOLOGY…
-
Dr. Penchovsky's award from the Bulgarian national contest, 2015
Awards for PostDocs
-
a young microbiologist national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
-
a young microbiologist in a national contest by the Foundation of Acad. Prof. Stephan Angeloff, 2023
PhD students' Awards
-
an award from the contest student of the year 2022 of Sofia University
-
an award from the national contest "Young and Energetic Scientists", 2021
our doctoral student Antoniya Georgieva won a first prize in the Ph.D. category…
Poster awards
-
Sofia Science Festival, May 15-16, 2021
see our acknowledged poster and research project RD-22-838/2020 by BMES…
-
an award from the Congress of Micro- biologists in Bulgaria with International Participation, Hisara 2018
Another awards
-
an award from the of the EWA 2022 Start-up Competition in Bulgaria, 2022
My doctoral student Antoniya Georgieva won a second place…
-
an award for teacher of the year for 125 School, Sofia, 2022
My doctoral student Georgi Miloshev won the teacher’s prize of the year…
-
an award for a contribution to the biology education of Sofia, 2023
My doctoral student Georgi Miloshev was awarded by the Bulgarian Ministry of Education and Science…
News and views on us
-
Homo Sciens
-
Robert's live interview for the Bulgarian National Radio about his upcoming talk on the Sofia Science Festival,
-
Our lab members' interview for Science_BG: video,
-
Homo Sciens
-
Nature Biotechnology
-
Nature Methods
-
ACS Synthetic Biology
-
RSC Chemistry World
-
Sofia University
Distinction for Prof. Dr. Robert Penchovsky from the Faculty of Biology…
-
YearBoook of Research Projects at Sofia University
Design and experimental validation of chimeric antisense oligo- nucleotides as antibacterial agents…
-
Magazine of Bulgarian Science
-
Sofia University
-
Bulgarian National Science Fund
-
Bulgarian Ministry of Education and Science
-
Magazine of Bulgarian Science
-
Interview with Prof. Draga Toncheva for the Bulgarian National Radio
-
Robert's interview for Science_BG: video,
-
Martina's short interview for Science_BG: video,
-
Robert's interview for Science_BG: podcast,
-
Robert's interview for the Bulgarian National Radio
-
News papers on us in Science_BG in Bulgarian, March, 2023
-
News on the main website of Sofia University in Bulgarian, March, 2023
-
News on the main website of Sofia University in English, April, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in Bulgarian, March, 2023
-
News on a young microbiologist national awards on website of the Institute of Microbiology, BAS, in English, March, 2023
-
BGlobal, in Bulgarian, July, 2023
A microbiologist replaces antibiotics when they do not work.…
-
Our recent paper is an editors' choice of the American Chemical Society. That is huge!
Ribozyme-based Molecular Circuit as Programmable Nanodevices
An important feature of molecular logic gates is the ability to generate signals that can control the activity of other molecular switches. The production of a diversity of highly-responsive oligonucleotide-sensing ribozymes would expand the complexity and efficiency of engineered nucleic acid circuits like those demonstrated previously.
To demonstrate a molecular cascading circuit, we employed construct YES-1 and a variation of construct YES-2 to create a simple molecular circuit (Fig. 1a). The sequence and length of YES-2 was altered in stem I to create a YES-2 variant that generates a new 21-nt 3` fragment on self-cleavage (Fig. 1a). This RNA fragment is complementary to the oligonucleotide binding site of YES-1, and therefore, it should activate the second ribozyme upon cleavage and dissociation from the first ribozyme in the circuit. This simple molecular signaling pathway was demonstrated using radiolabeled YES-1 with various combinations of unlabeled YES-2 and the oligonucleotides DNA-1 and DNA-2. Although DNA-1 triggers YES-1 cleavage as demonstrated here, no cleavage is observed when both ribozymes are simultaneously incubated in the absence of DNA effector (Fig. 1b). This demonstrates that the YES-1 ribozyme is not activated when its RNA-1 signal oligonucleotide remains attached to the YES-2 variant ribozyme. In contrast, the addition of DNA-2 induces YES-1 cleavage only when the YES-2 variant ribozyme is present. Furthermore, the kinetics of YES-1 function as part of the complete signaling pathway indicate a lag phase that we interpret to be caused by the time required to release RNA-1 from the YES-2 variant upon activation by DNA-2 (Fig. 1c). These findings are consistent with the design of the ribozyme signaling pathway wherein an activator of the first ribozyme in the series triggers the release of an activator of the second ribozyme. Although simple in design, our findings suggest that a variety of more complex molecular circuitry could be constructed wherein oligonucleotide triggers could be used to carry out a variety of logic-based ribozyme functions.
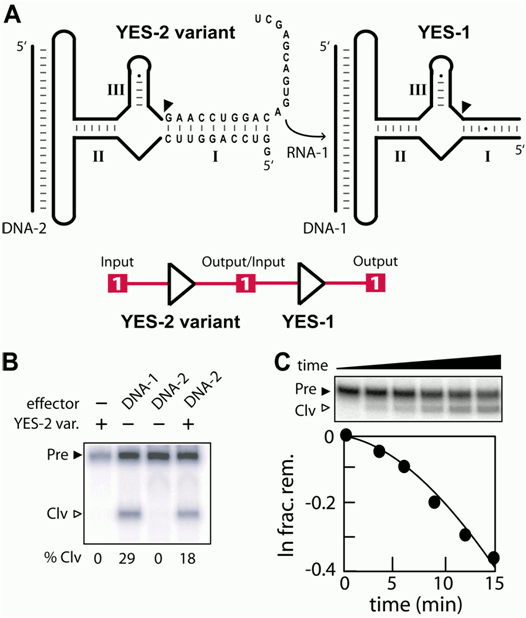
Figure 1. Ribozyme-based molecular cascading circuit using YES-1 and a variant of YES-2. (A) Nucleotides of the YES-2 variant RNA that differ from YES-2 are depicted. Upon activation of YES-2 variant by effector DNA-2, the 3` cleavaged fragment (RNA-1) is released and serves as an effector for YES-1 activation. Schematic presentations of the logic circuit of the two YES gates are depicted. (B) Assay depicting function of a ribozyme signaling pathway. YES-1 RNAs are radiolabeled in all lanes. (C) Kinetic analysis of YES-1 self-cleavage in the presence of YES-2 variant and its effector DNA-2. Details are as described in B.
References: 1. Robert Penchovsky & Ronald R. Breaker - Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes – 2005, Nature Biotechnology, 10870156, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 43,5. 2. Robert Penchovsky - Engineering integrated digital circuits with allosteric ribozymes for scaling up molecular computation and diagnostics – 2012, ACS Synth Biol, 21615063, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 5,382 3. Robert Penchovsky's Short Biography - ACS Synth Biol - 2012, 21615063, Q1 (Biochemistry, Genetics and Molecular Biology), IF – 5,382