|
|
 |
Login: | |
Sitemap: | Search: |
Robert Penchovsky's Website
RSwitch results
Dear Visitor, you can see the results below:
Information about the selected riboswitch displayed below
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
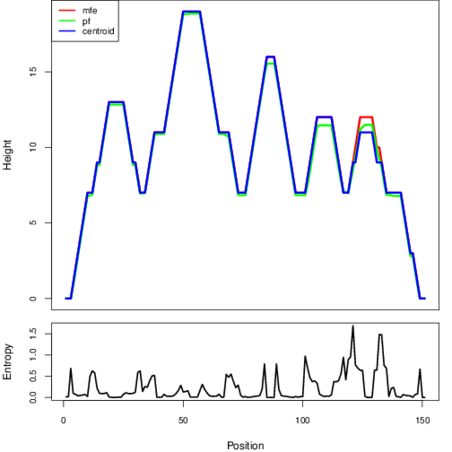
The plot represents the thermodynamic ensemble of the RNA structure of the Acinetobacter baymannii ATCC 17978, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
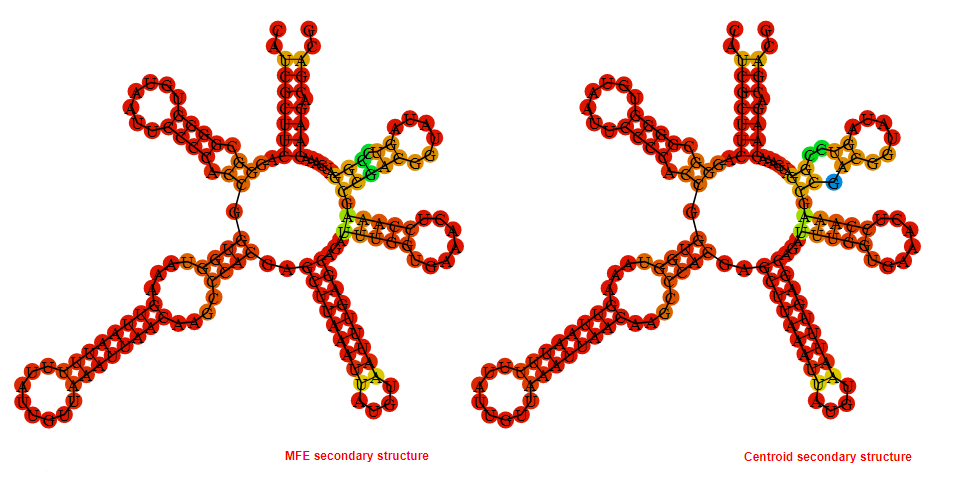
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Acinetobacter baymannii ATCC 17978, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
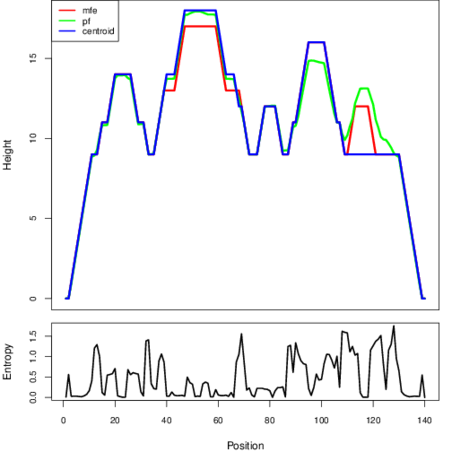
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis str Western North America, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
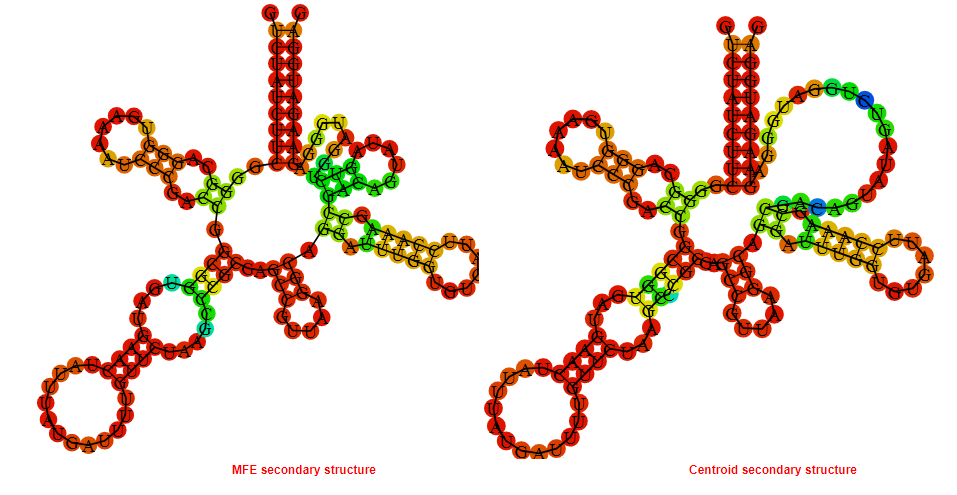
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis str Western North America, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
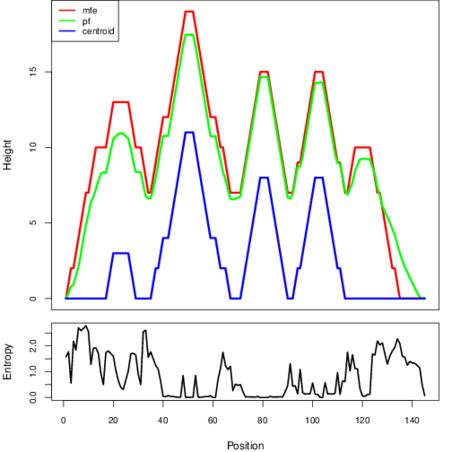
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis_1, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
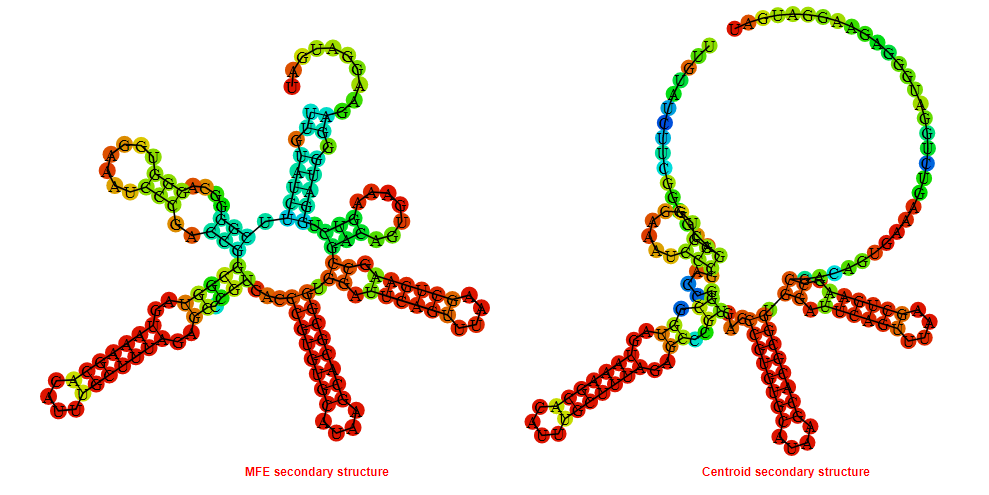
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis_1, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
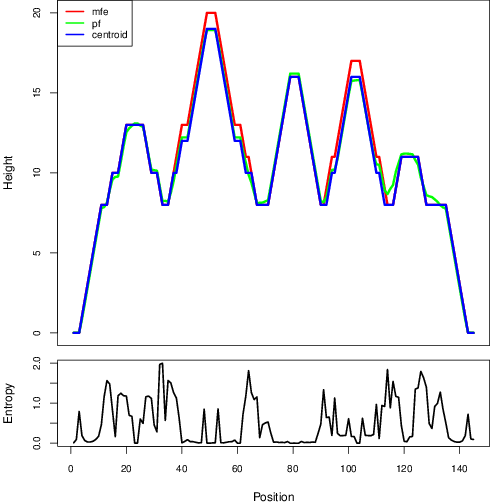
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis_2, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
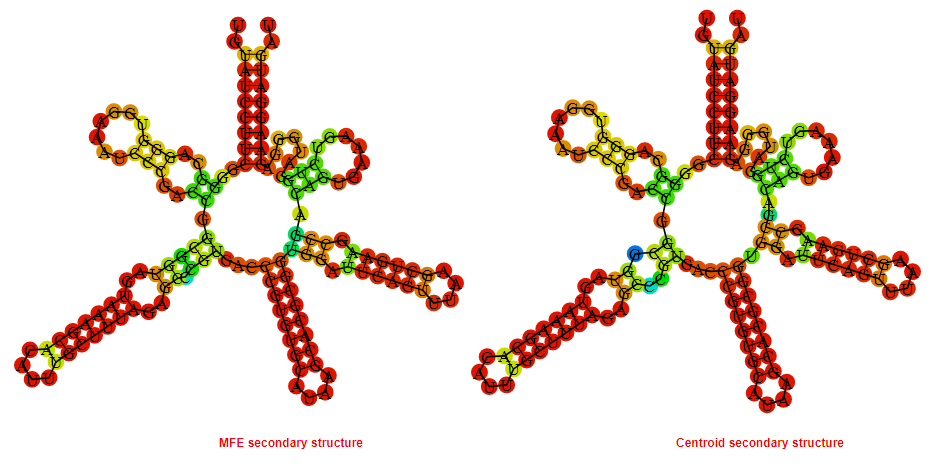
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis_2, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
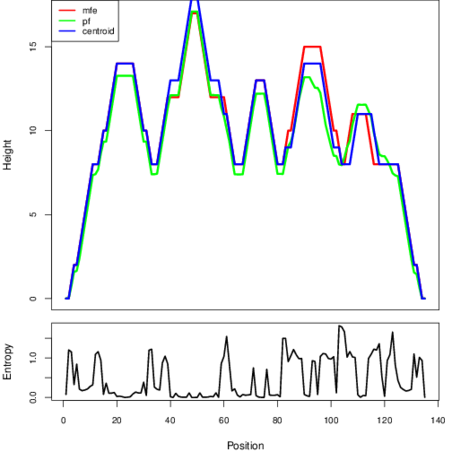
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis_3, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
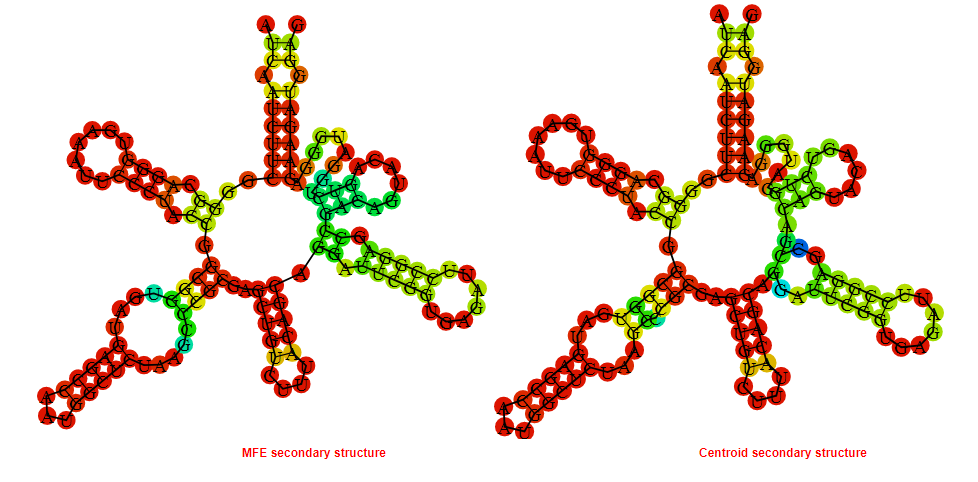
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis_3, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
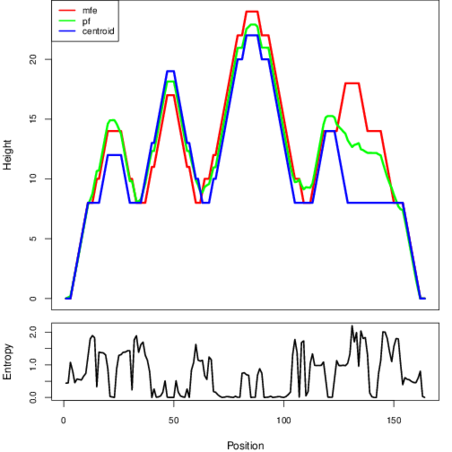
The plot represents the thermodynamic ensemble of the RNA structure of the Brucella canis ATCC 23365, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
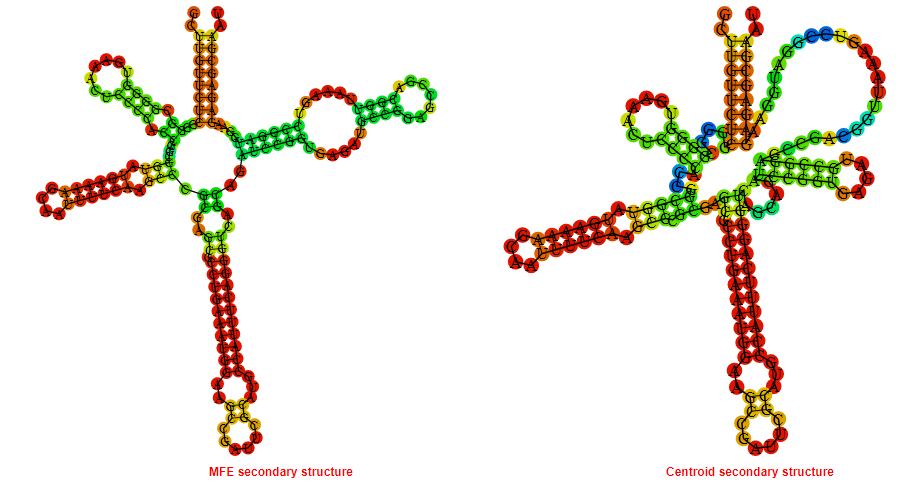
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Brucella canis ATCC 23365, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
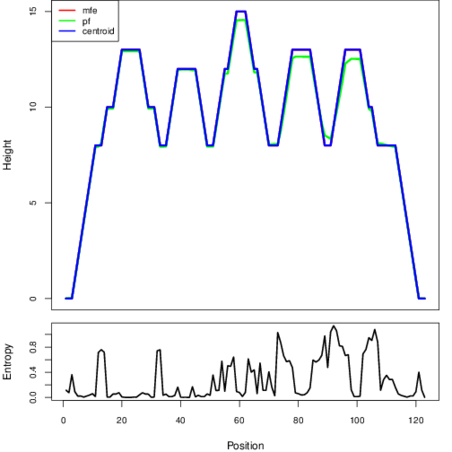
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum C str_Eklund , FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
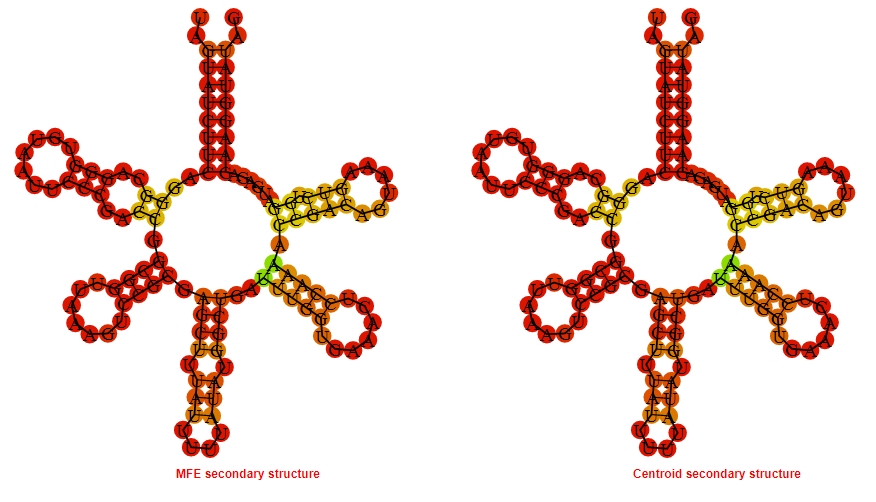
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum C str_Eklund , FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
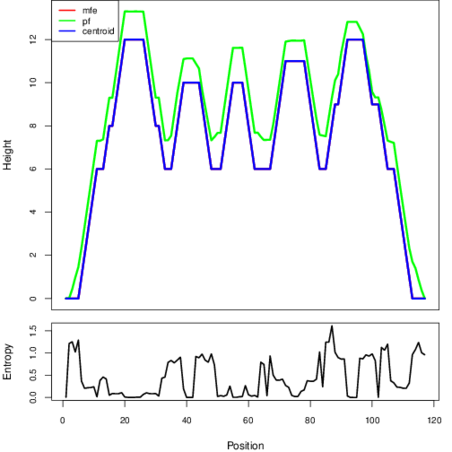
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum A2 str_Kyoto, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
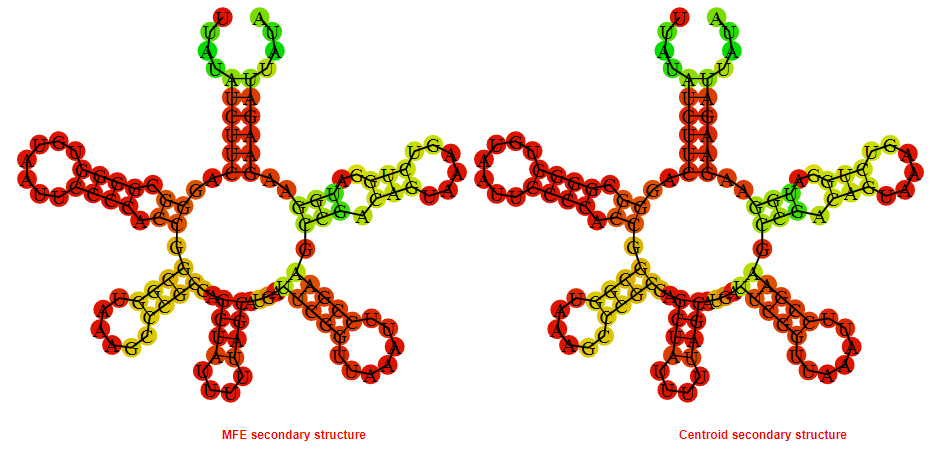
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum A2 str_Kyoto, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
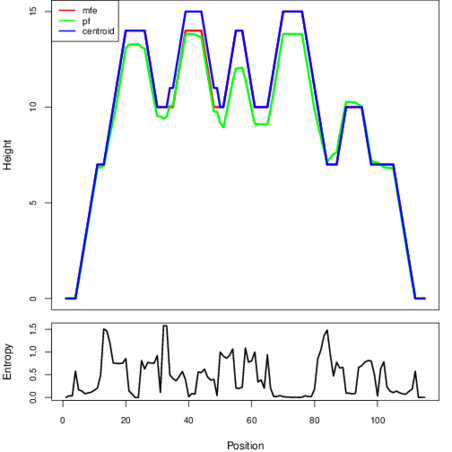
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum E4 str_BoNT_E_BL5262, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
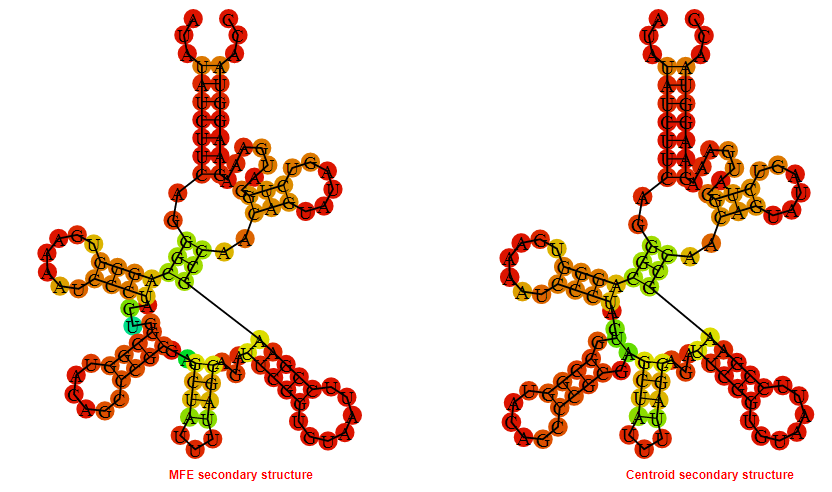
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum E4 str_BoNT_E_BL5262, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
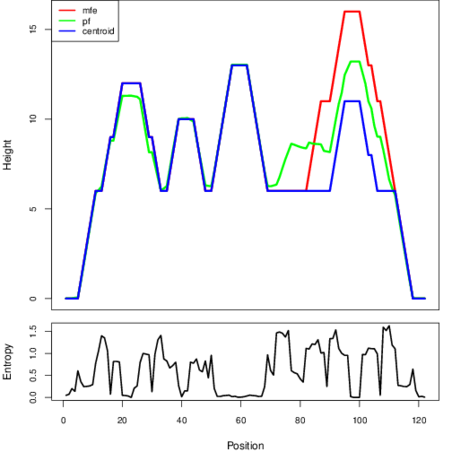
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
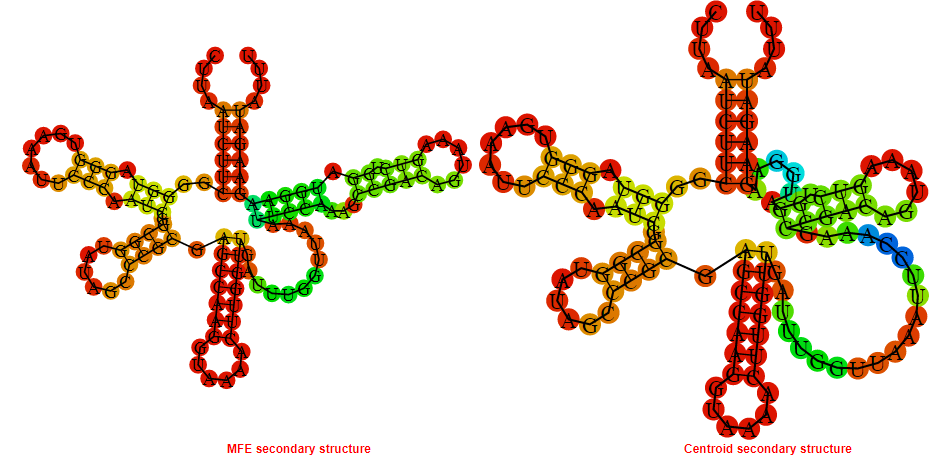
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
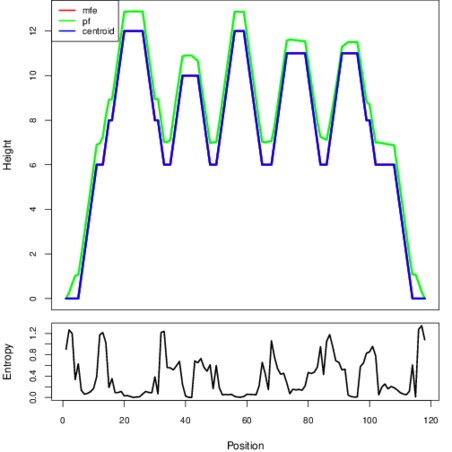
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile QCD32g58, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
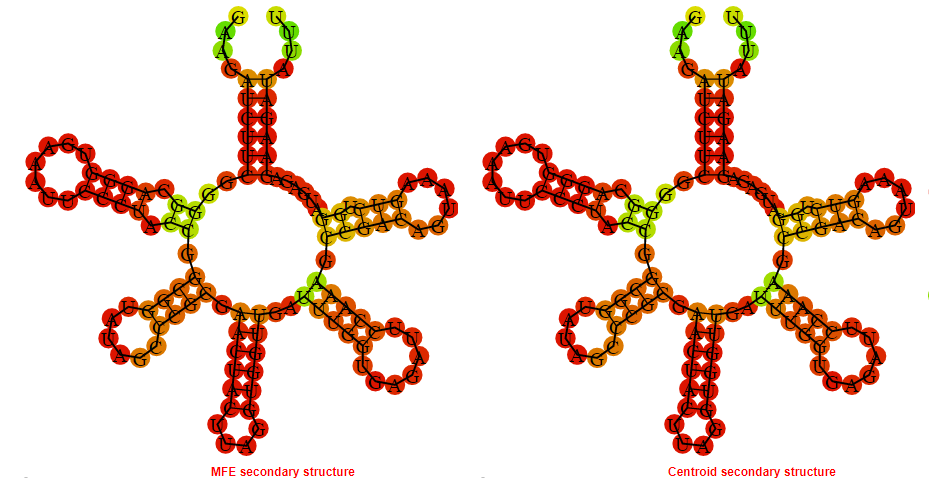
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile QCD32g58, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
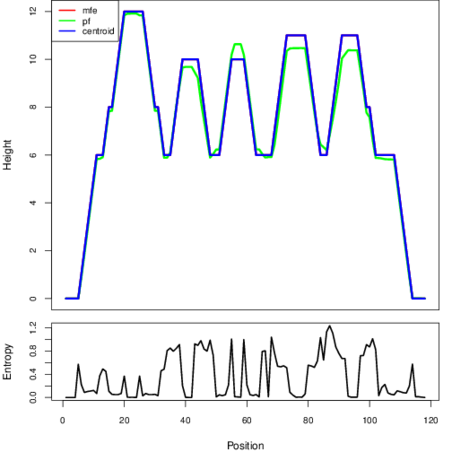
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium_perfringens CPE str_F4969, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
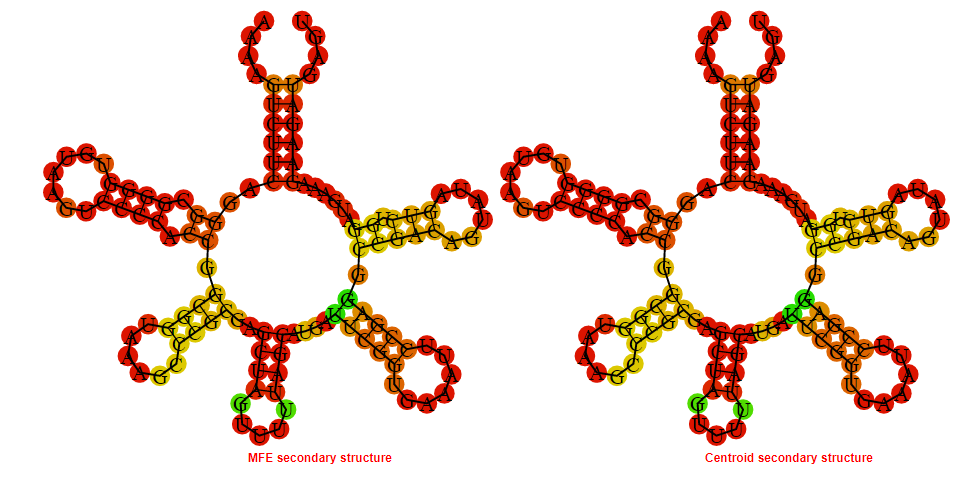
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium_perfringens CPE str_F4969, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
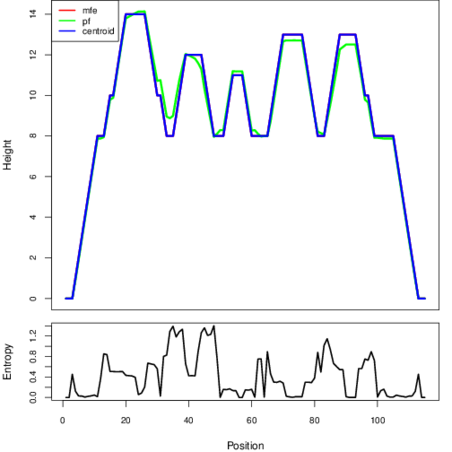
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani_E88, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
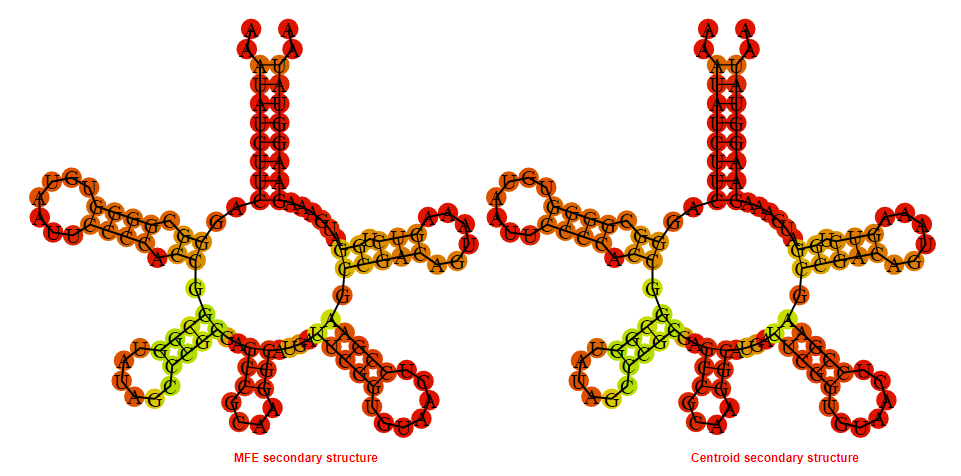
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani_E88, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
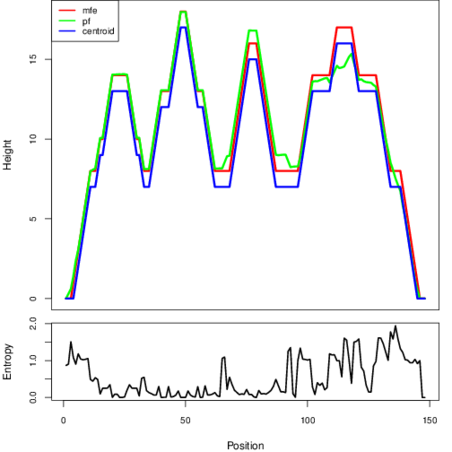
The plot represents the thermodynamic ensemble of the RNA structure of the Escherichia coli, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
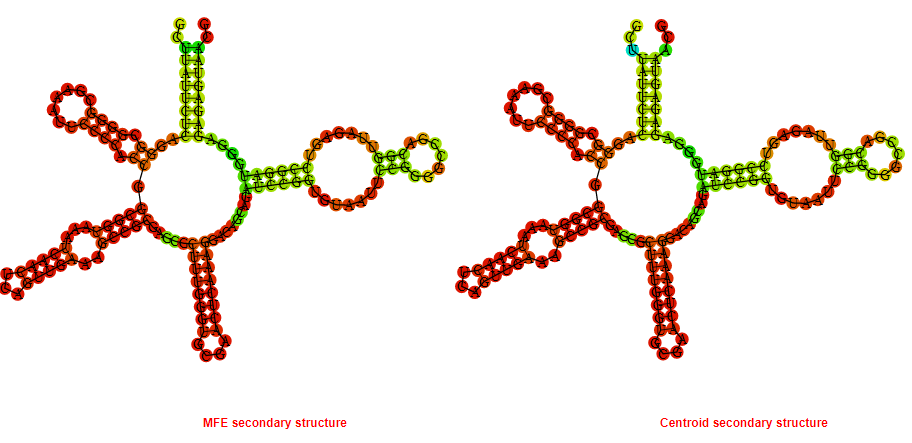
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Escherichia coli, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
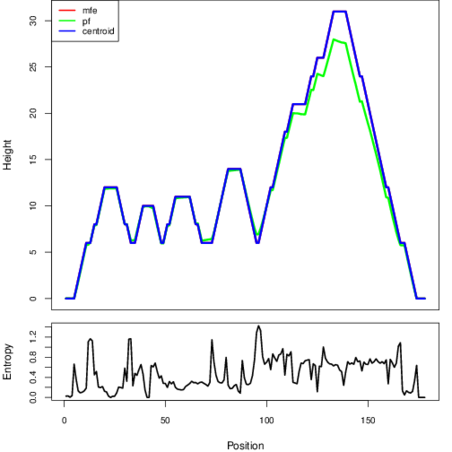
The plot represents the thermodynamic ensemble of the RNA structure of the Haemophilus influenzae_22121, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
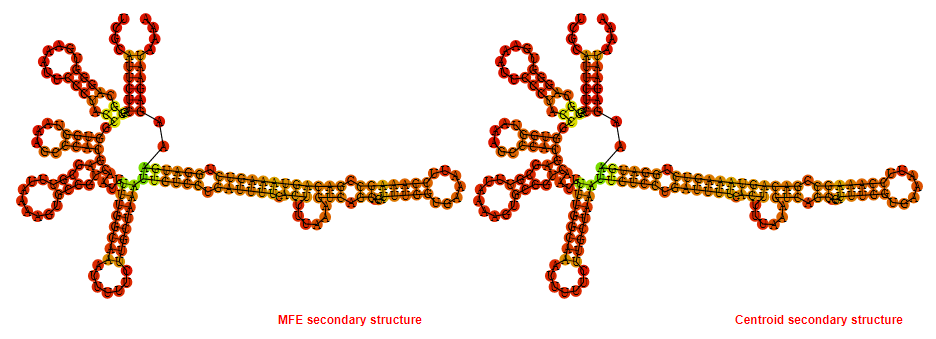
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Haemophilus influenzae_22121, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
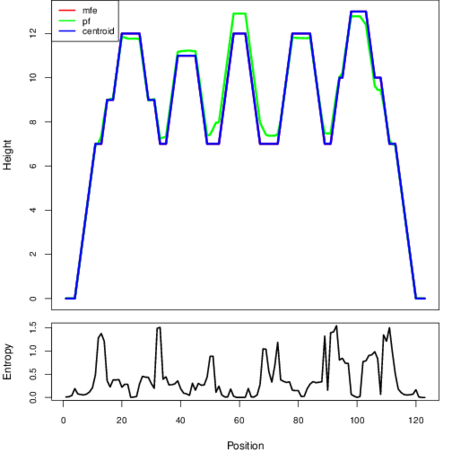
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
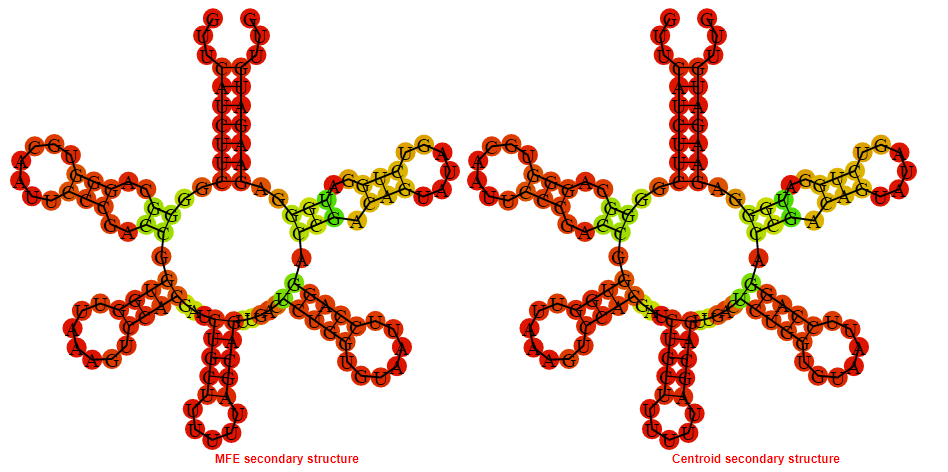
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
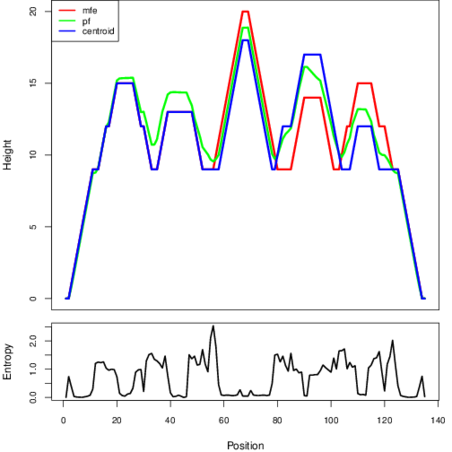
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus subsp_aureus_COL, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
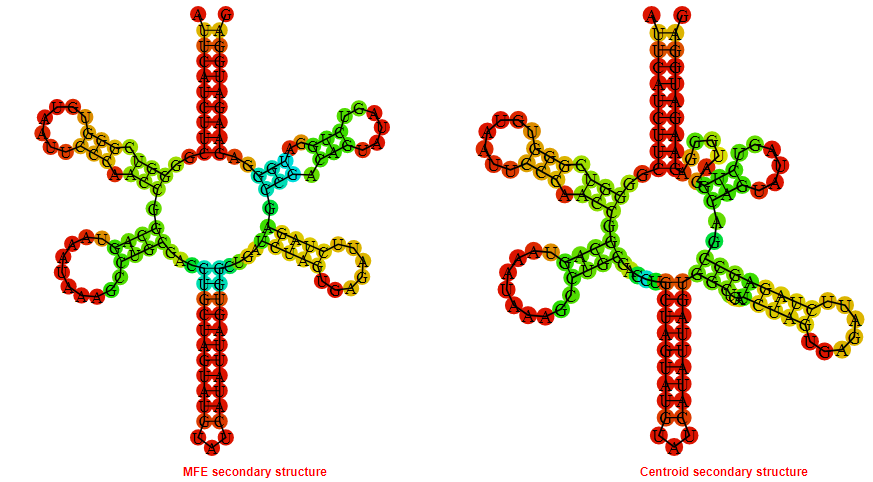
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus subsp_aureus_COL, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
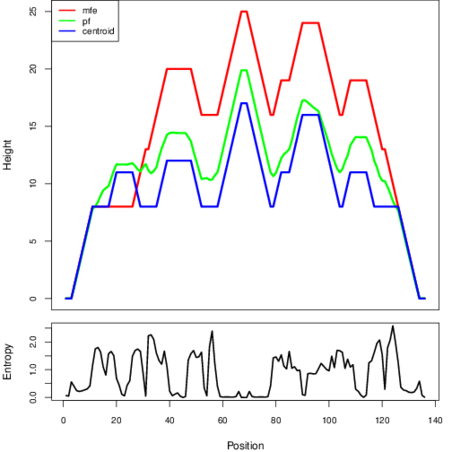
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus subsp_aureus_JH9, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
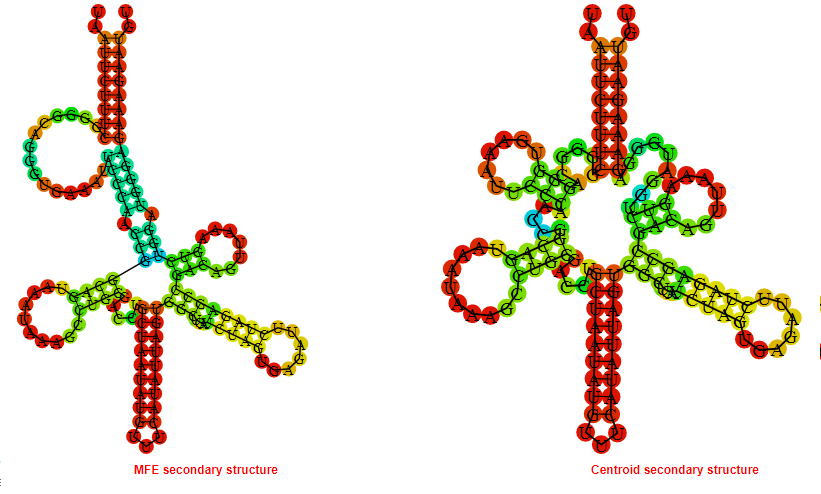
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus subsp_aureus_JH9, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
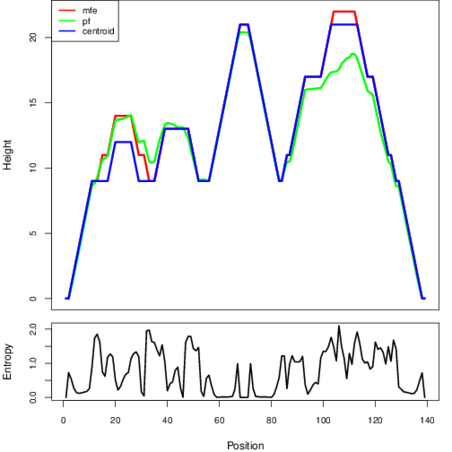
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis_1, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
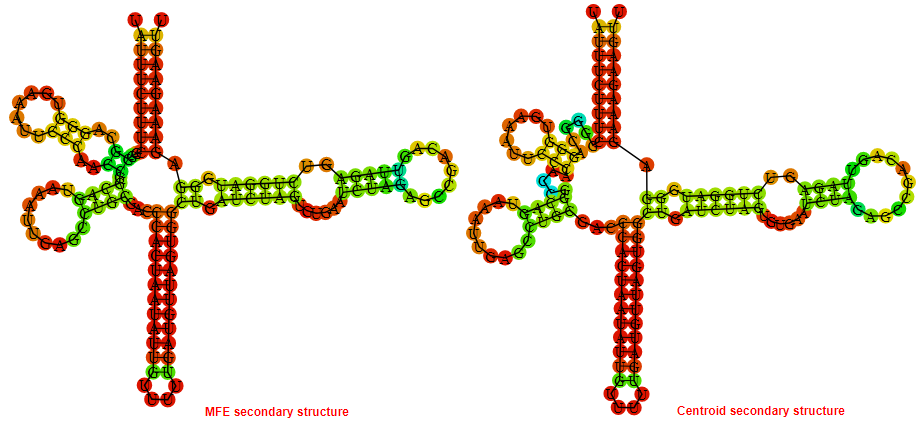
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis_1, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
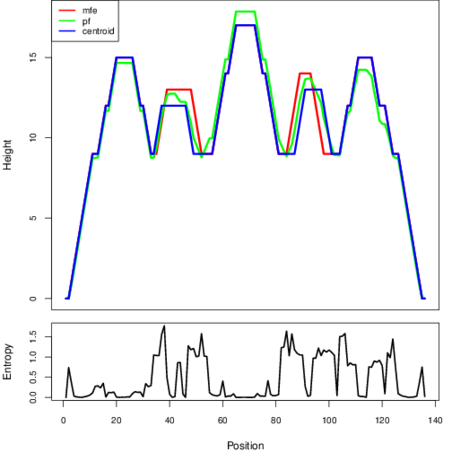
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis_2, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
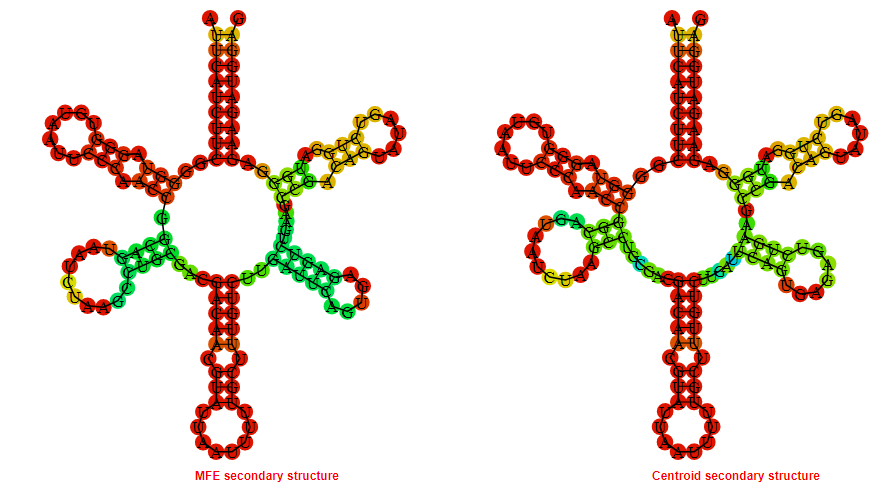
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis_2, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
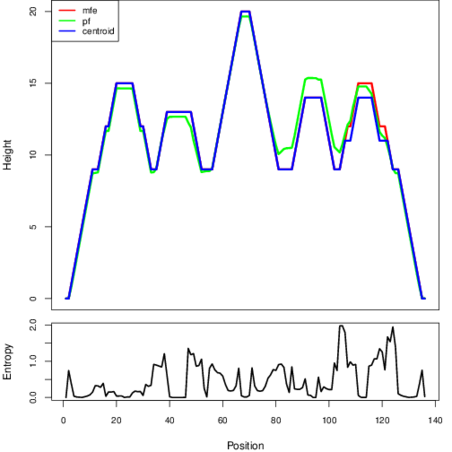
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus subsp saprophyticus ATCC_15305, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
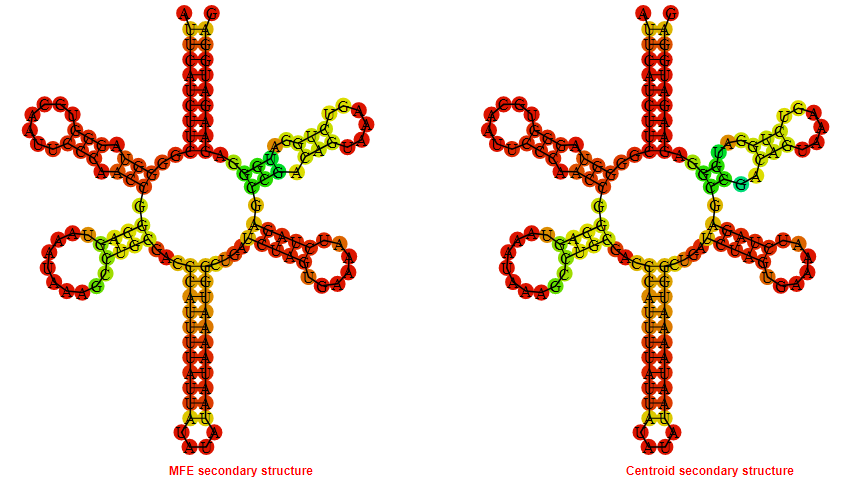
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus subsp saprophyticus ATCC_15305, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
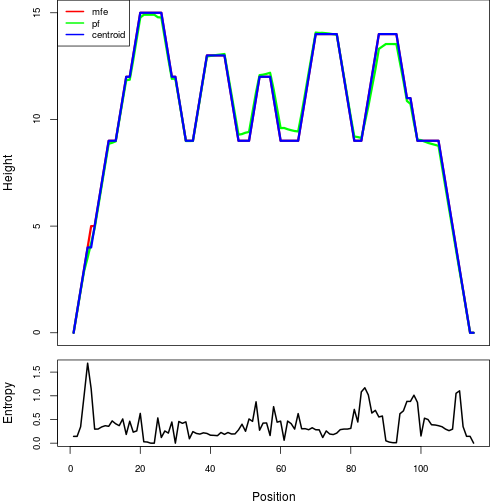
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus agalactiae A909, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
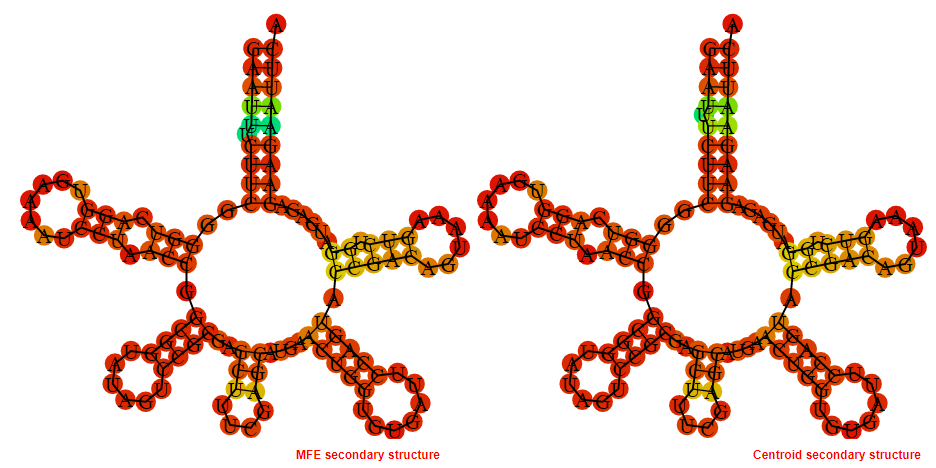
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus agalactiae A909, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
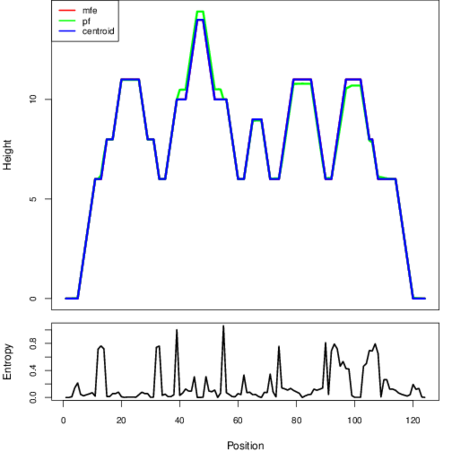
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pyogenes MGAS10750, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
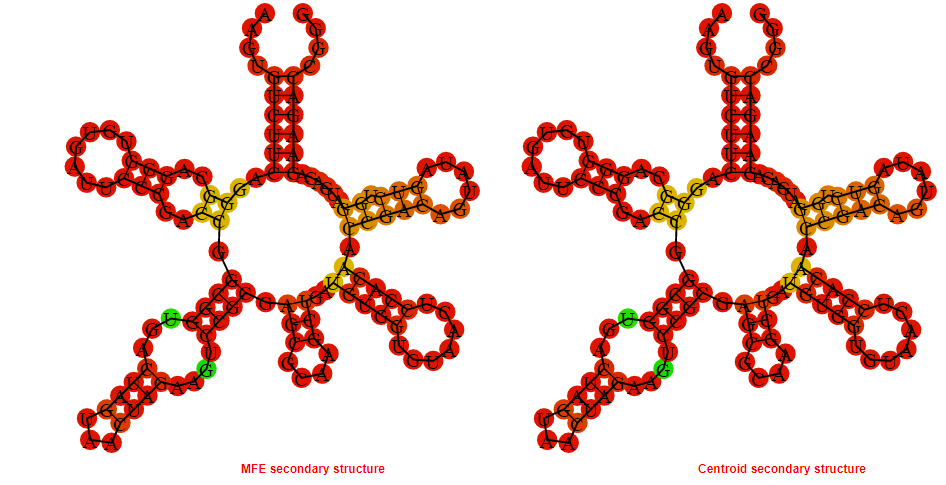
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pyogenes MGAS10750, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
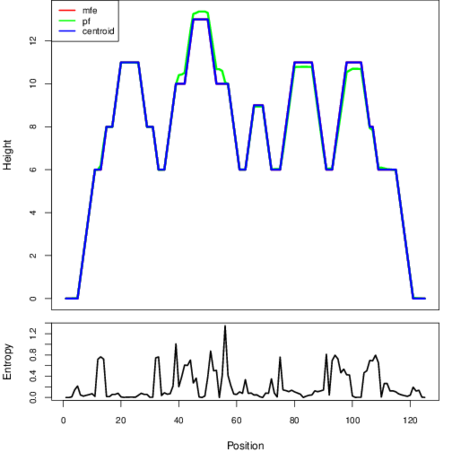
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pyogenes MGAS2092, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
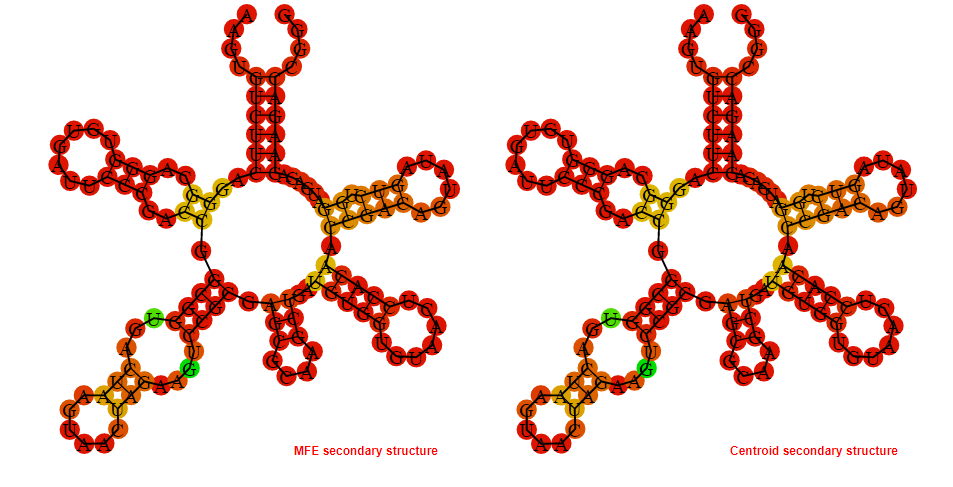
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pyogenes MGAS2092, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
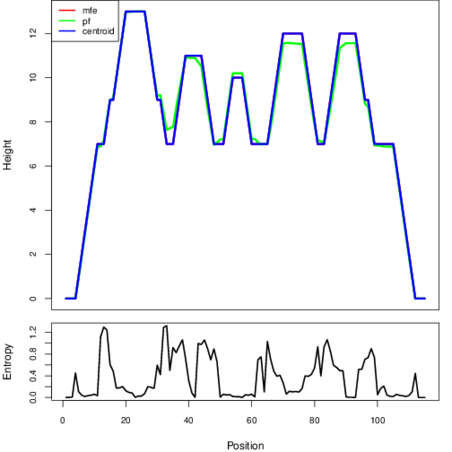
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pneumoniae JJA, FMN type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
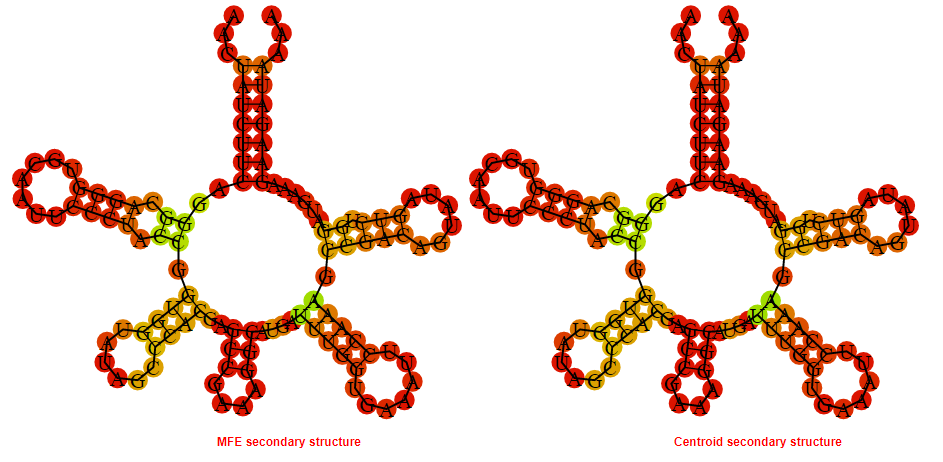
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pneumoniae JJA, FMN type switch.
Multiple alignment of the FMN type switches
Below is presented the sequences alignment of all the FMN type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the FMN type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the FMN type switches involved in.

Multi_resistance strains for the FMN type switches
The table displays known human pathogenic bacteria bearing the FMN type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
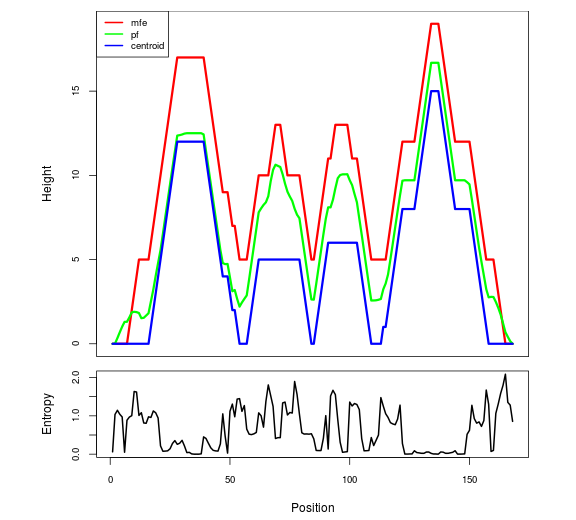
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis 1, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
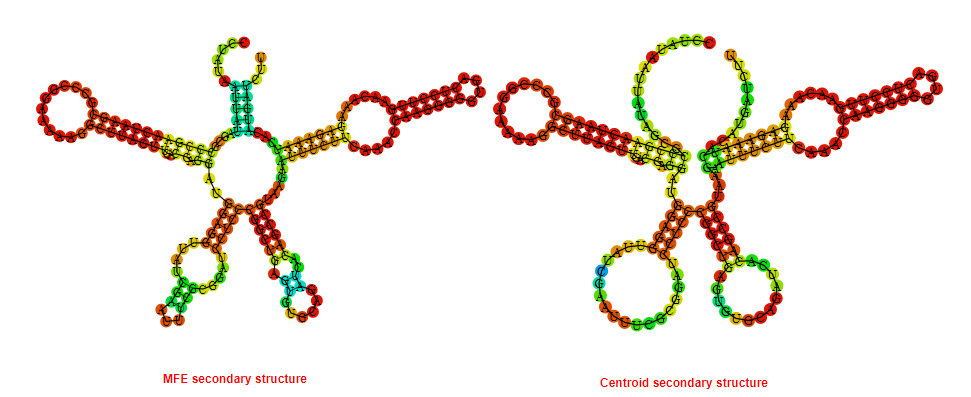
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis 1, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.
Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
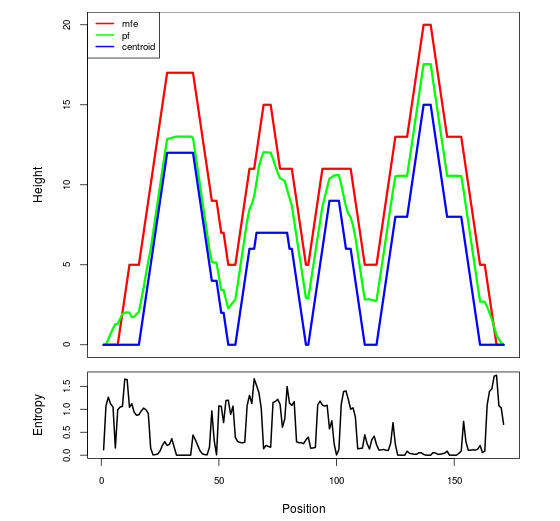
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis subsp spizizenii ATCC_6633_1, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
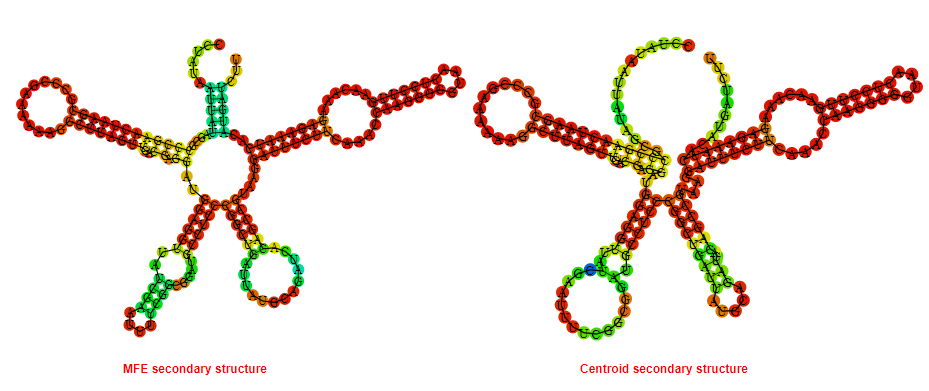
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis subsp spizizenii ATCC_6633_1, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
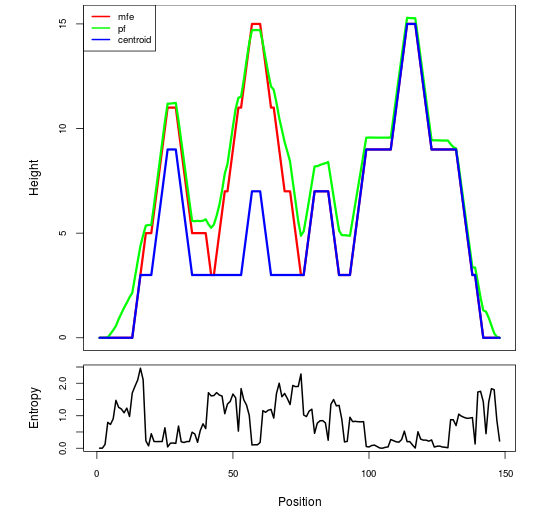
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis_V583, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
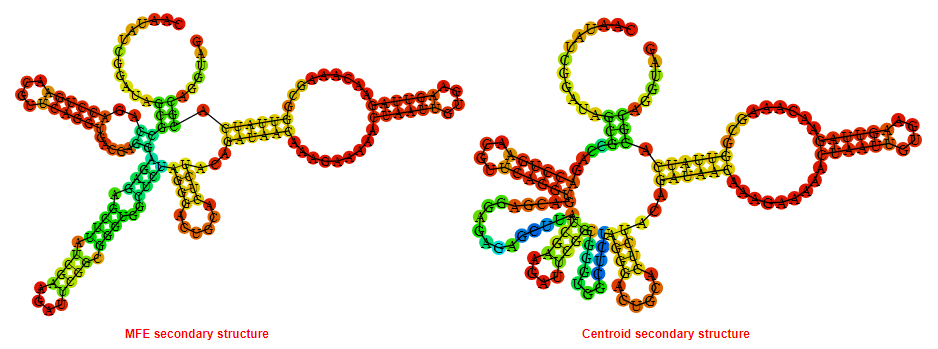
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis_V583, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
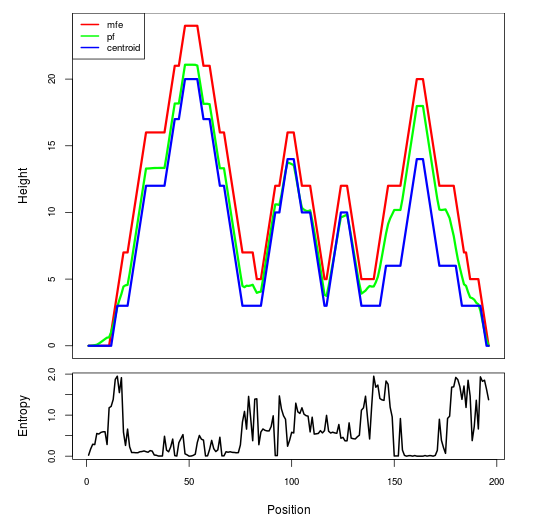
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria moncytogenes str 4b F2365_1, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
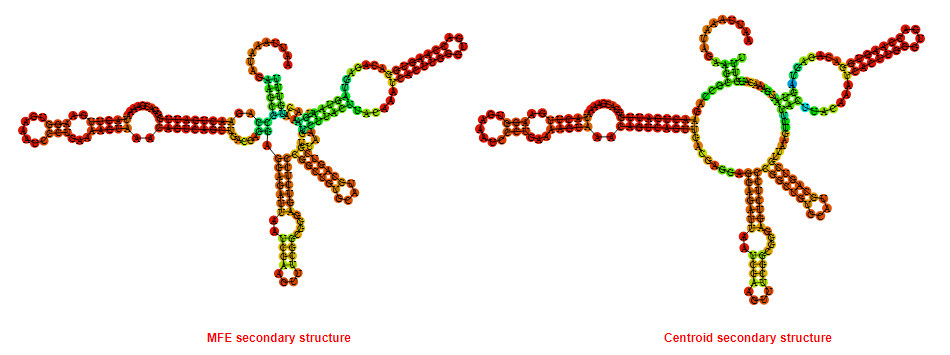
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria moncytogenes str 4b F2365_1, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
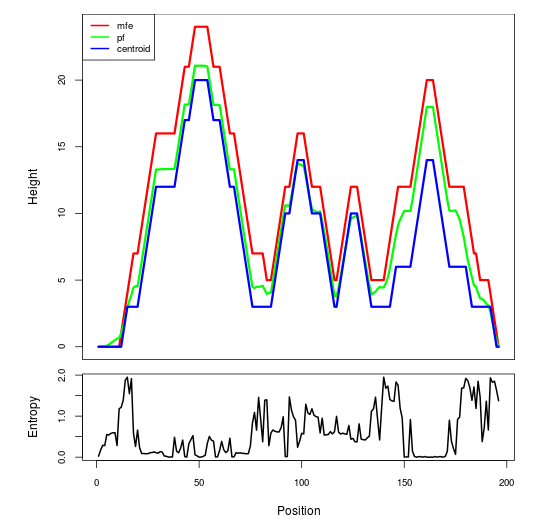
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes FSL_N3_165, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
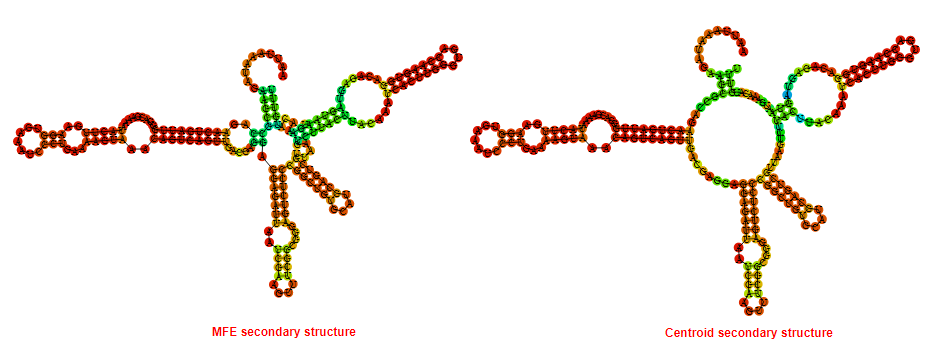
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes FSL_N3_165, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
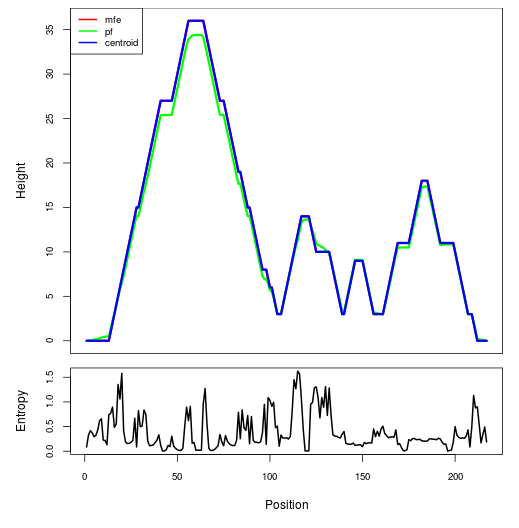
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
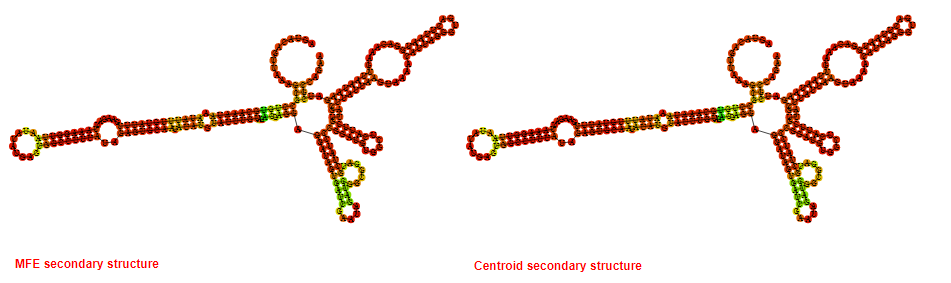
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
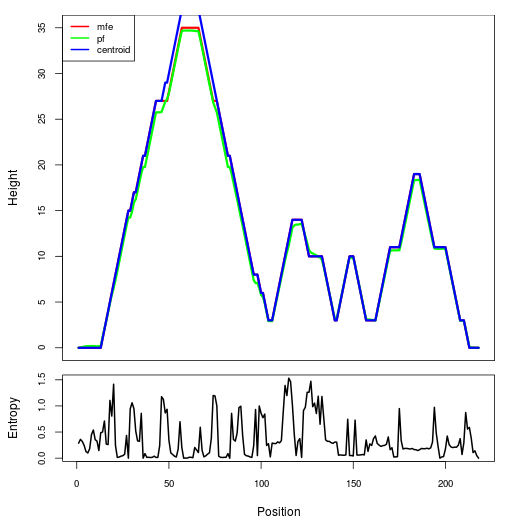
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
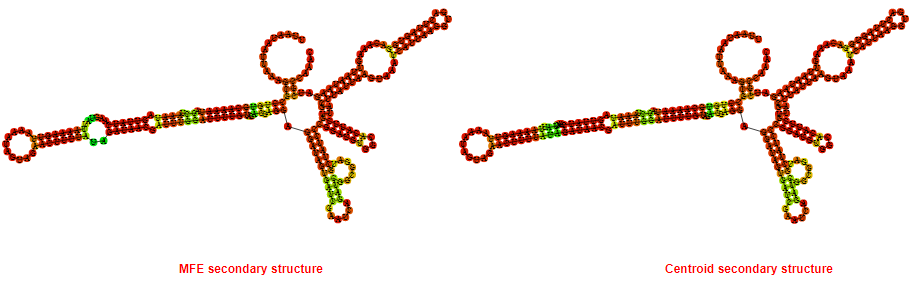
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
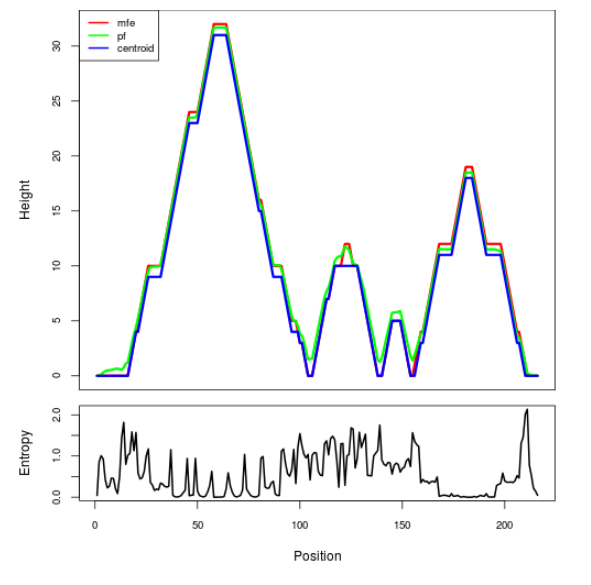
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcuss saprophyticus ATCC_15305, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
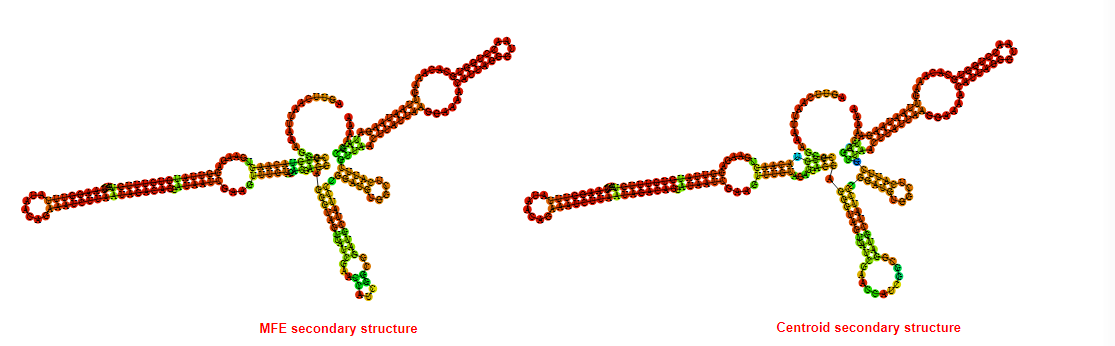
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcuss saprophyticus ATCC_15305, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
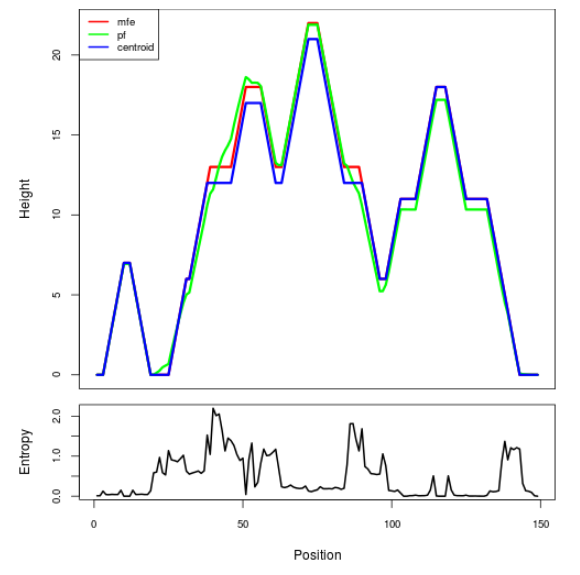
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
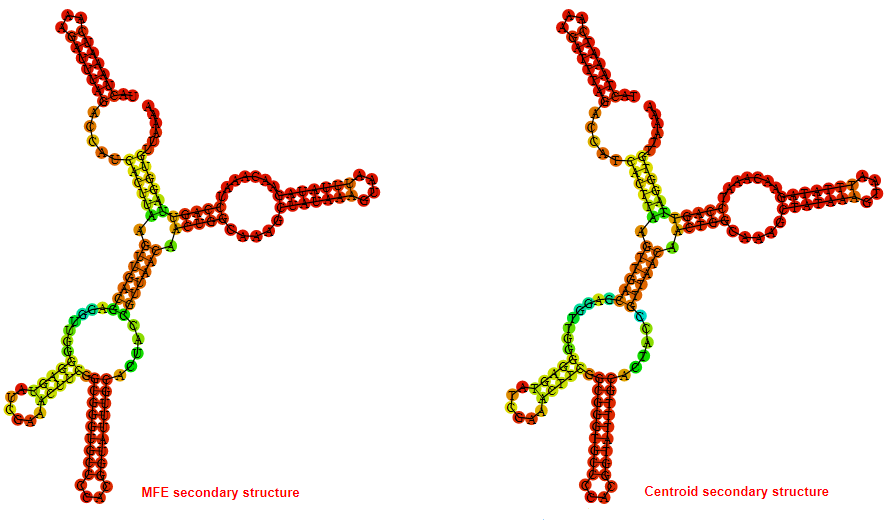
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure

The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
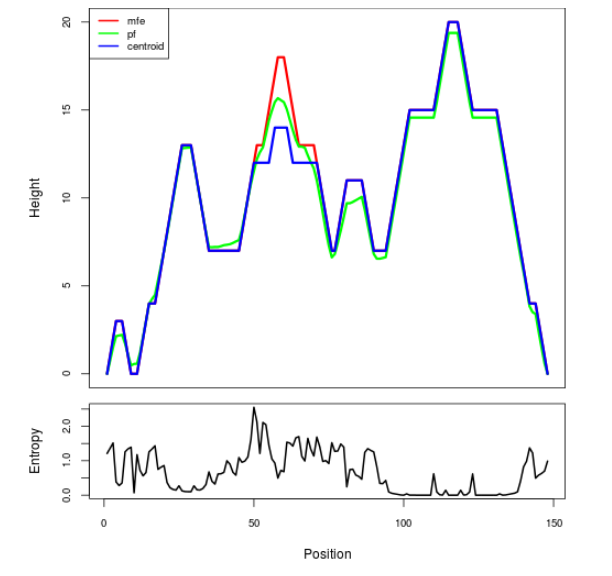
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
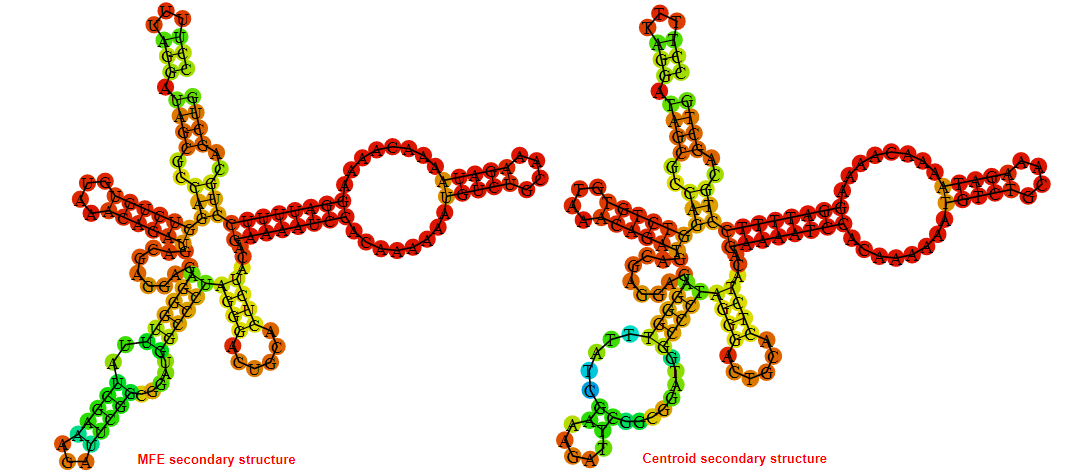
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
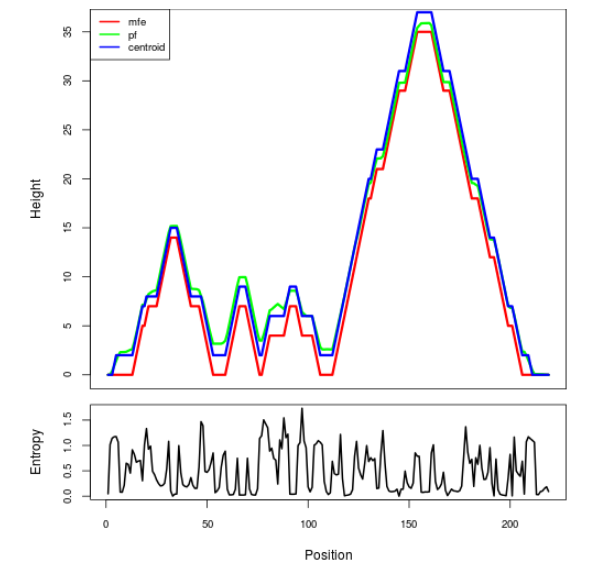
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
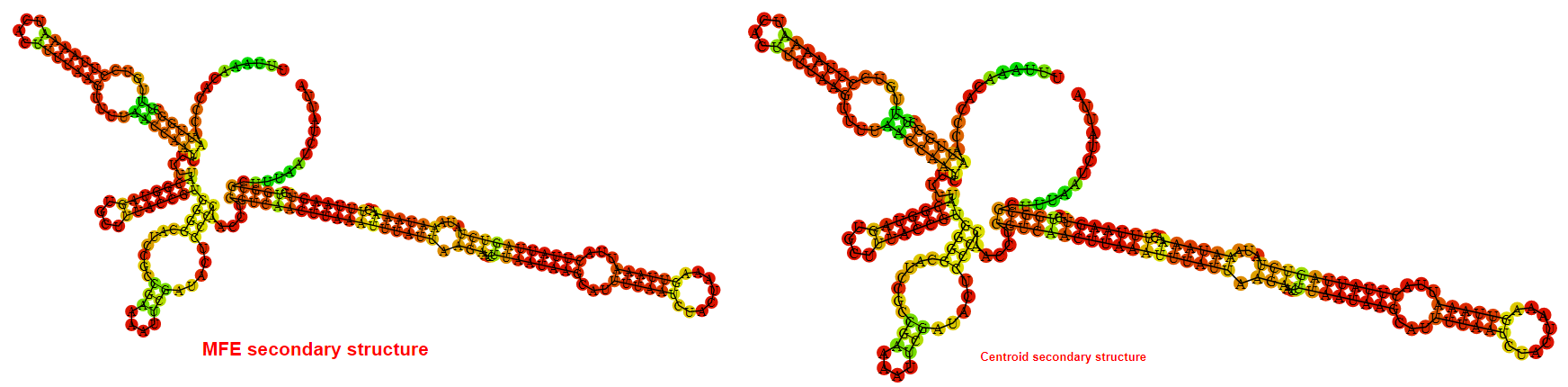
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
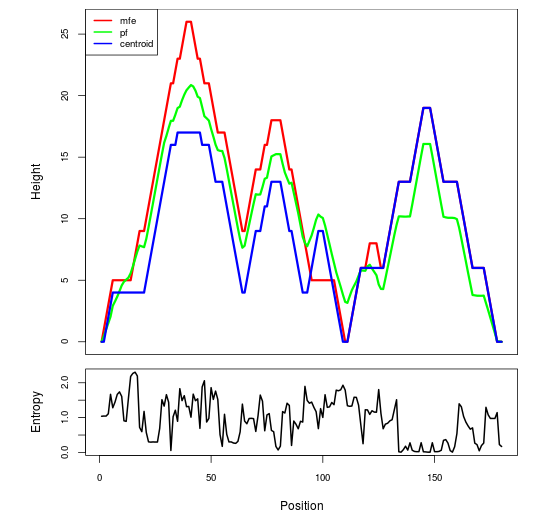
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile, GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
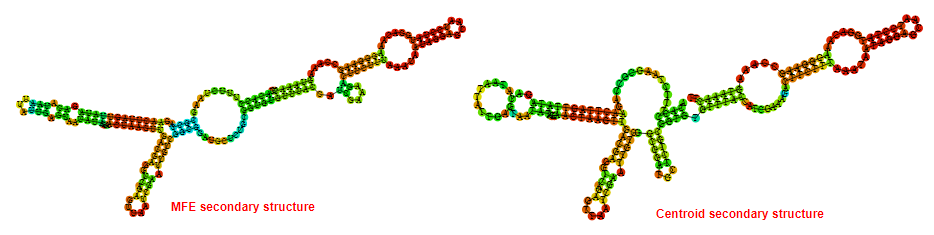
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile, GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
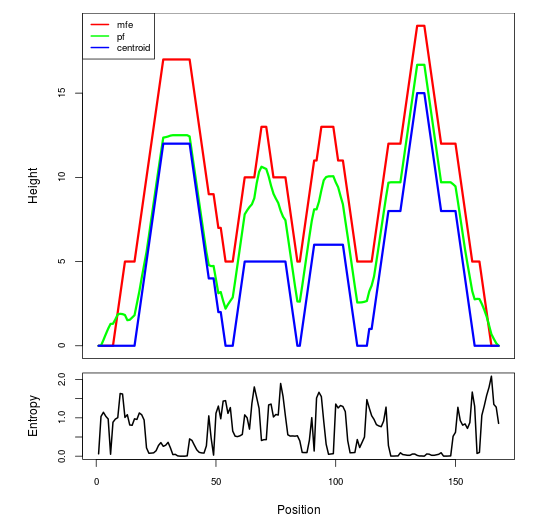
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis , GlmS type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
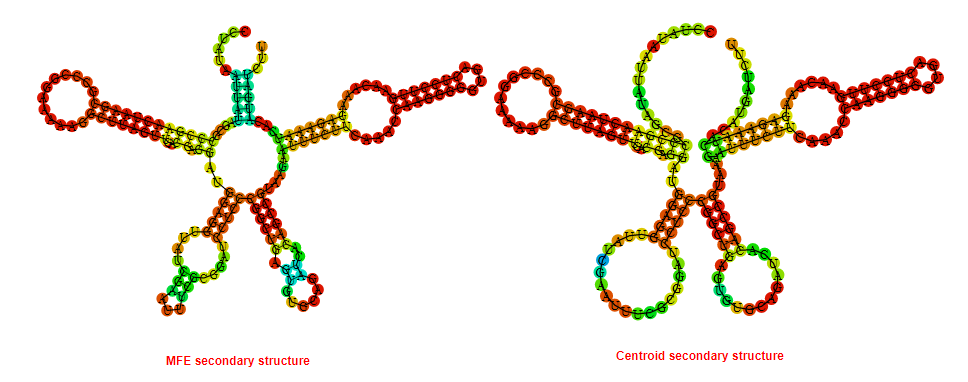
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis , GlmS type switch.
Multiple alignment of the GlmS type switches
Below is presented the sequences alignment of all the GlmS type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the GlmS type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the GlmS type switches involved in.

Multi_resistance strains for the GlmS type switches
The table displays known human pathogenic bacteria bearing the GlmS type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
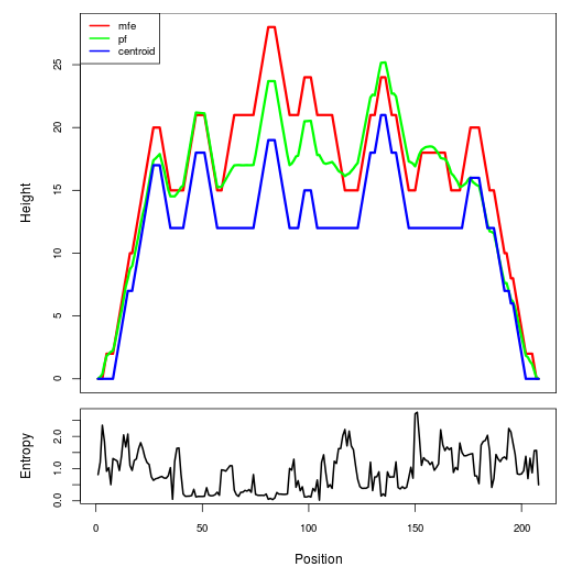
The plot represents the thermodynamic ensemble of the RNA structure of the Acinetobacter baumannii , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
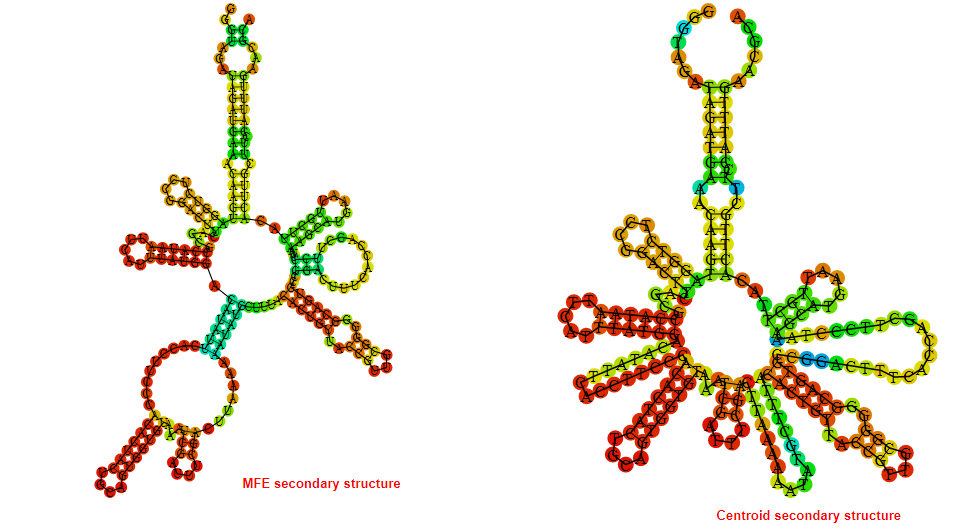
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Acinetobacter baumannii , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.
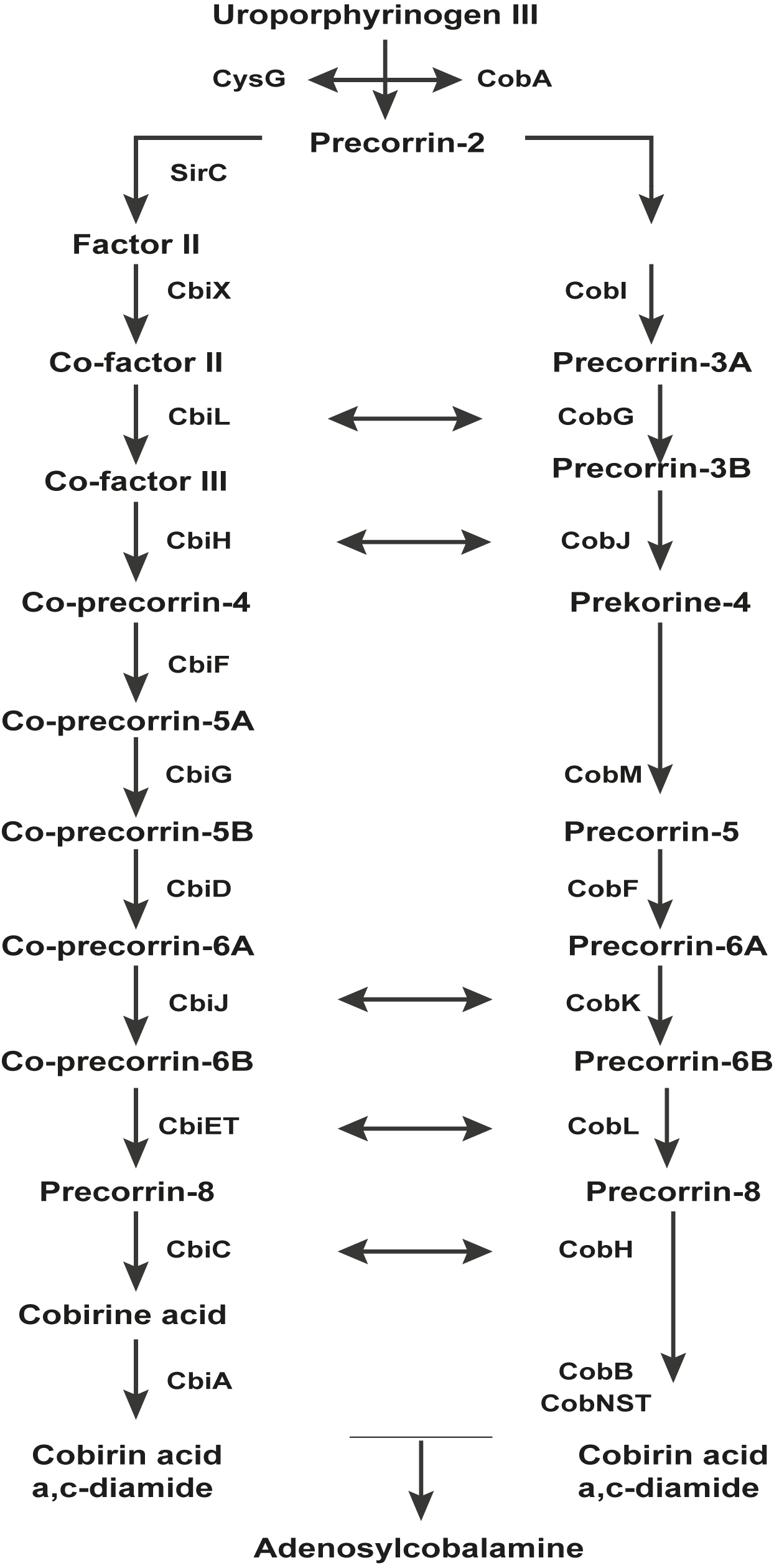
Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
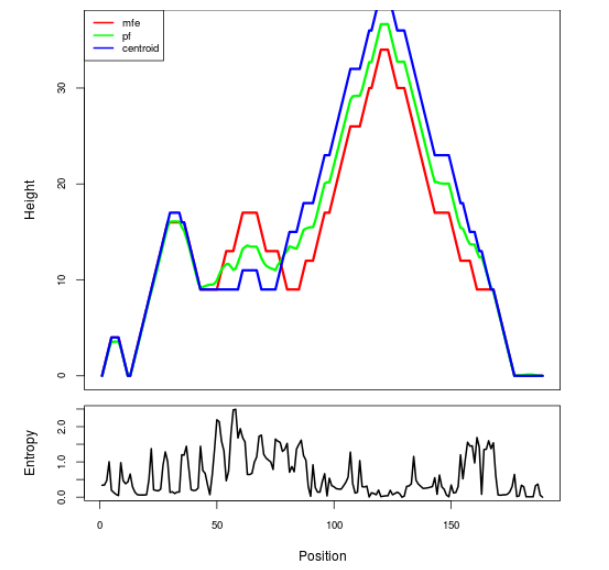
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
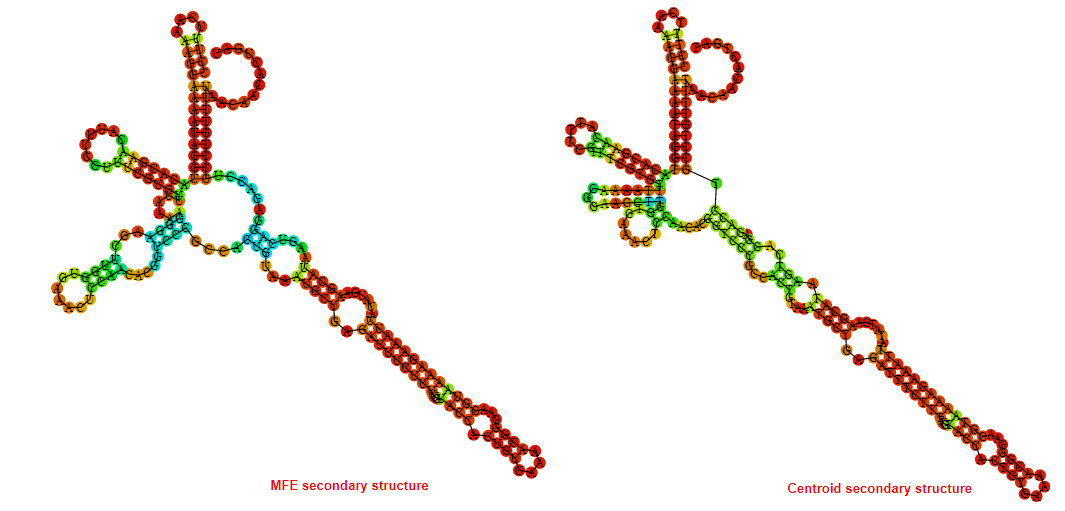
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
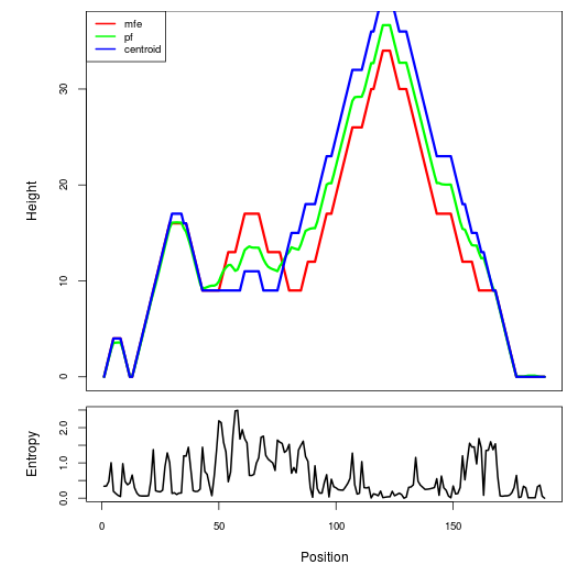
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus cereus , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
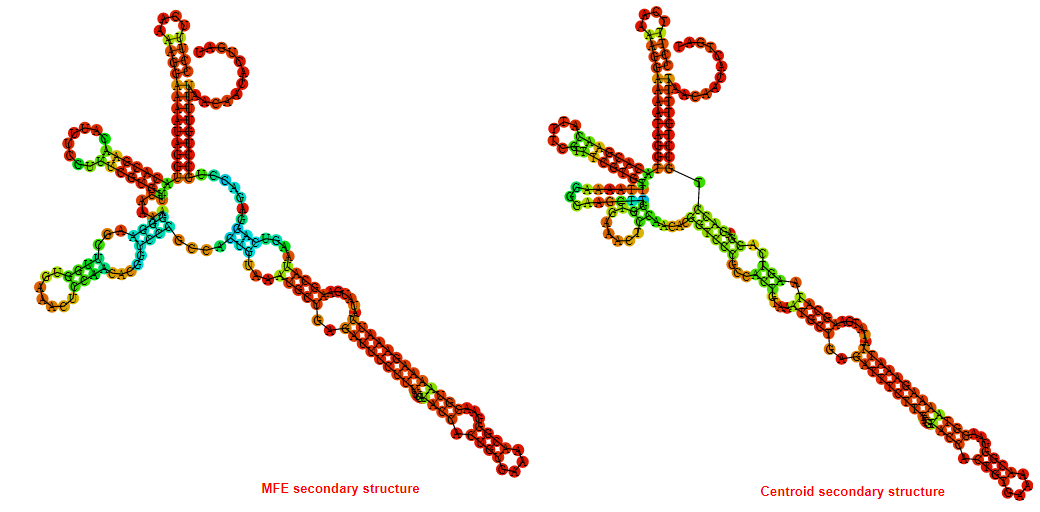
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus cereus , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
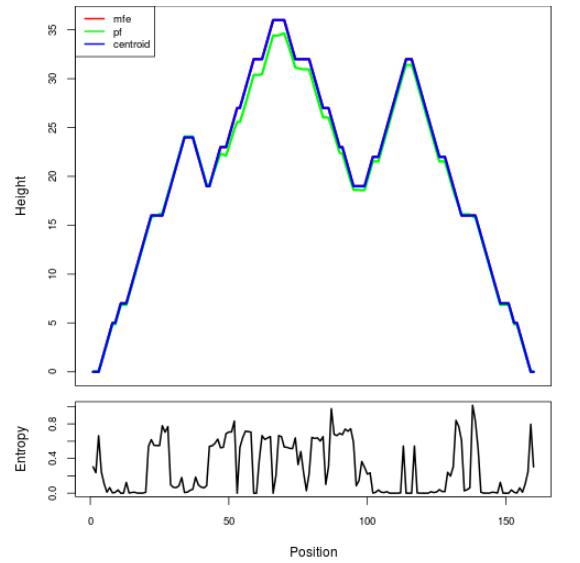
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis strain Tohama I, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
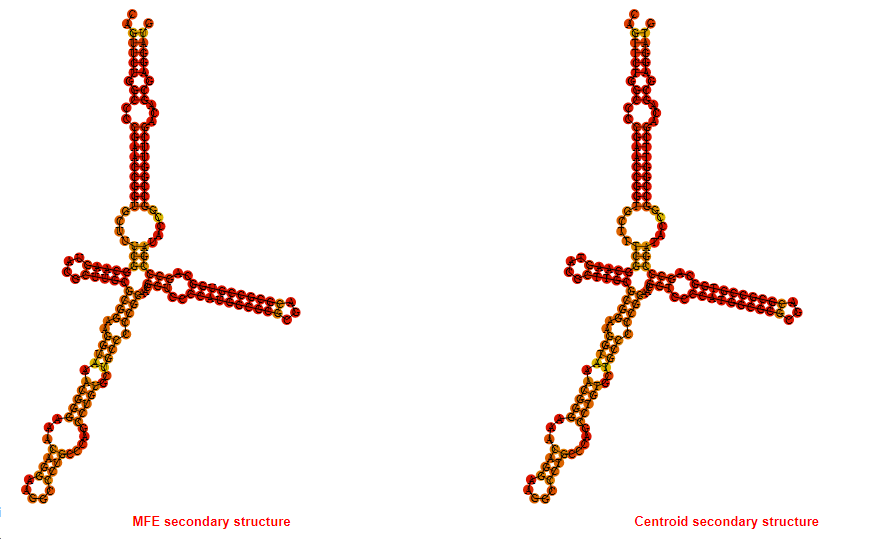
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis strain Tohama I, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
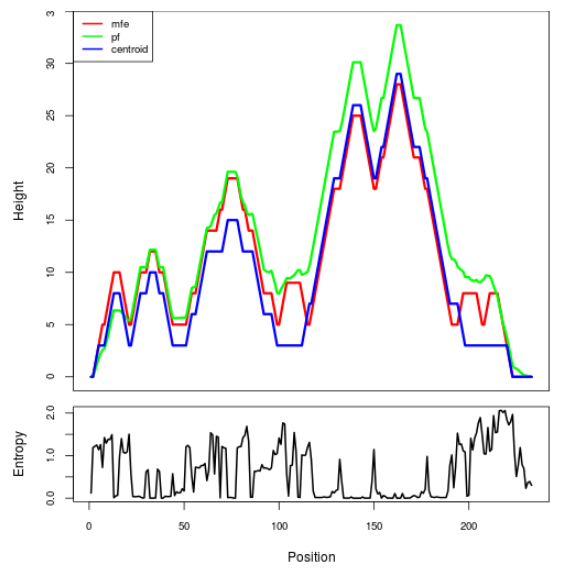
The plot represents the thermodynamic ensemble of the RNA structure of the Brucella melitensis biovar Abortus , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
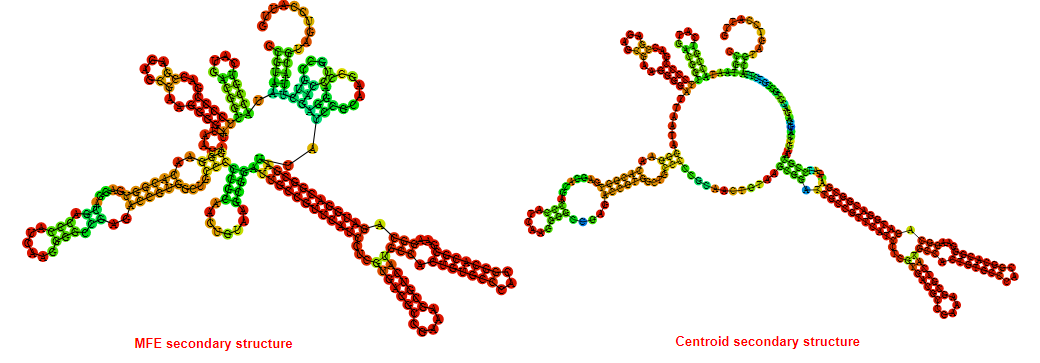
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Brucella melitensis biovar Abortus , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
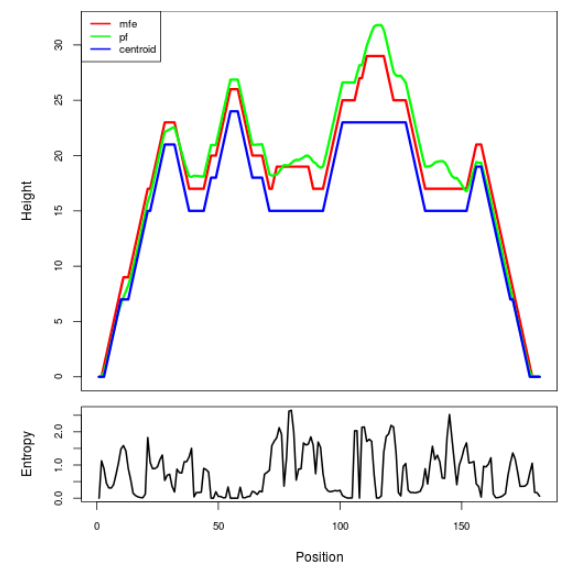
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum A str. ATCC 3502 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
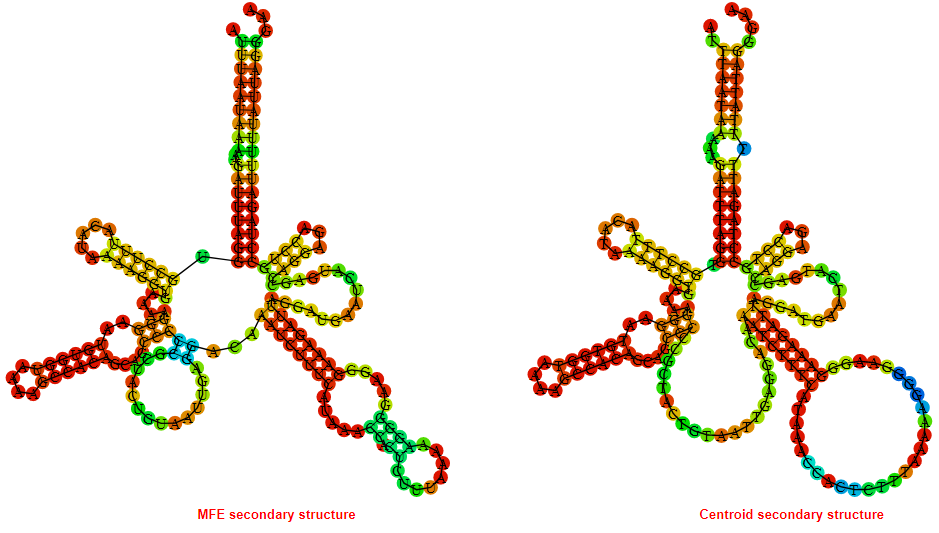
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum A str. ATCC 3502 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
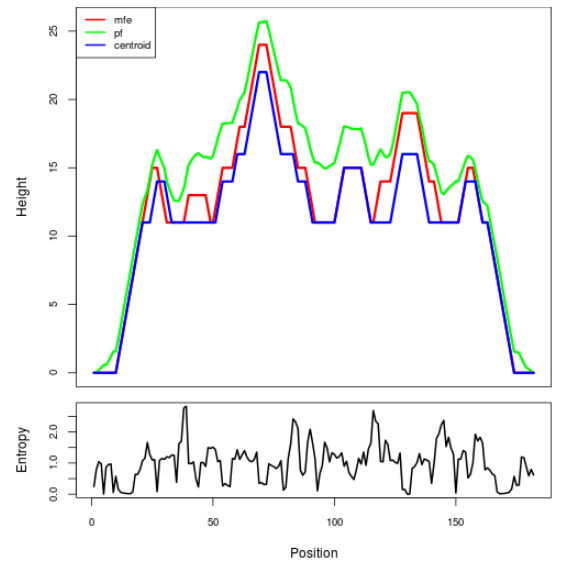
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
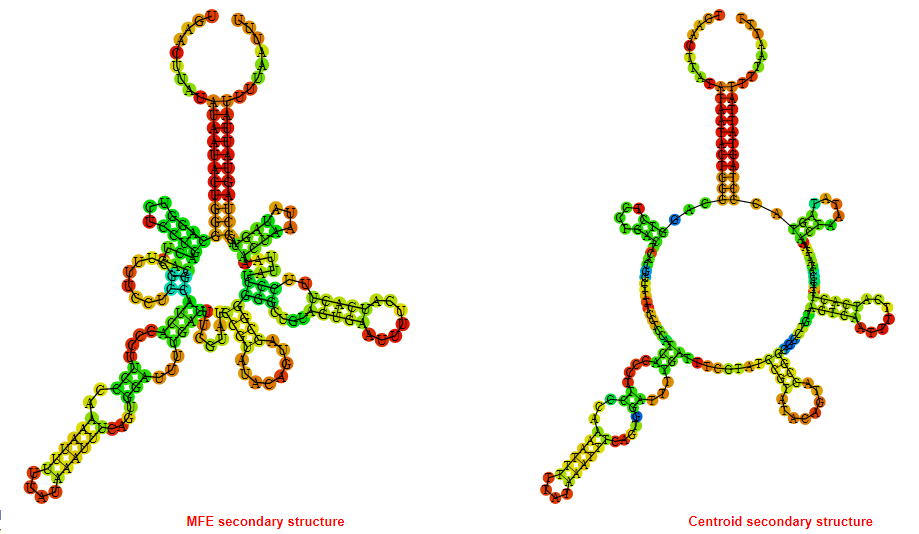
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
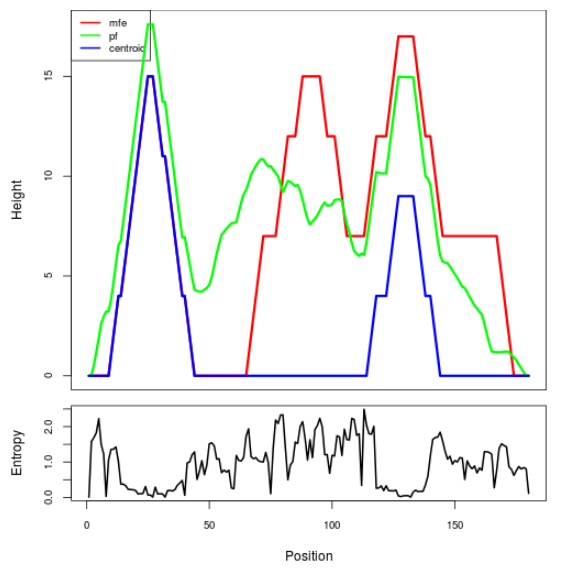
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens str.13 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
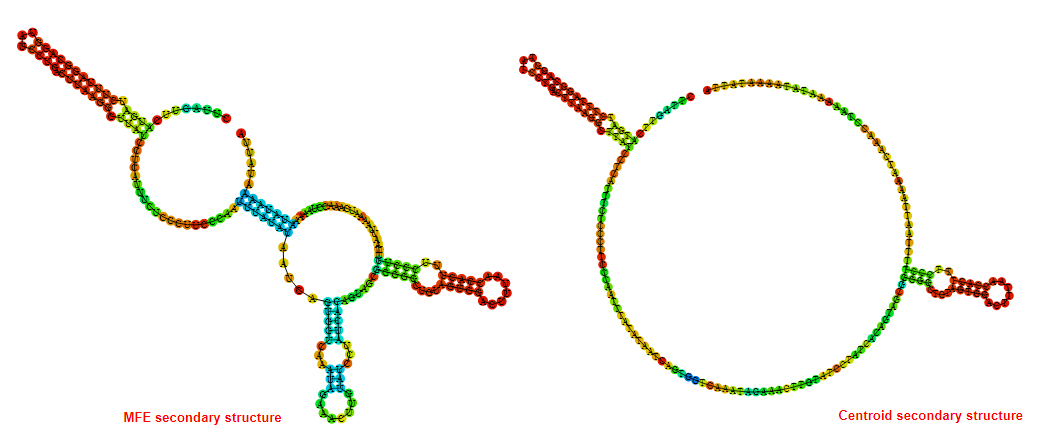
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens str.13 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
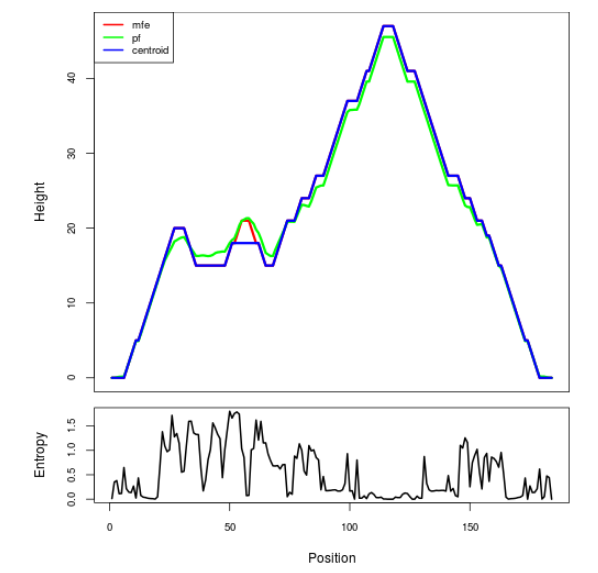
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani E88, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
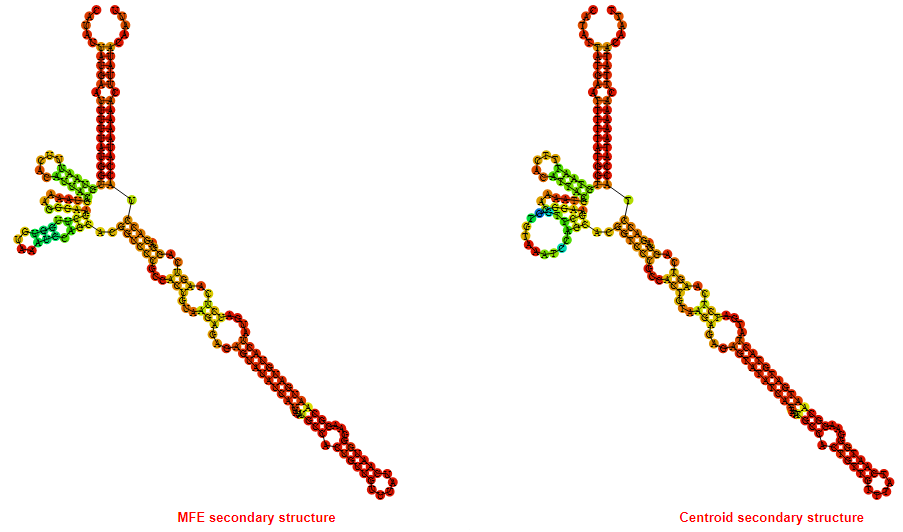
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani E88, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
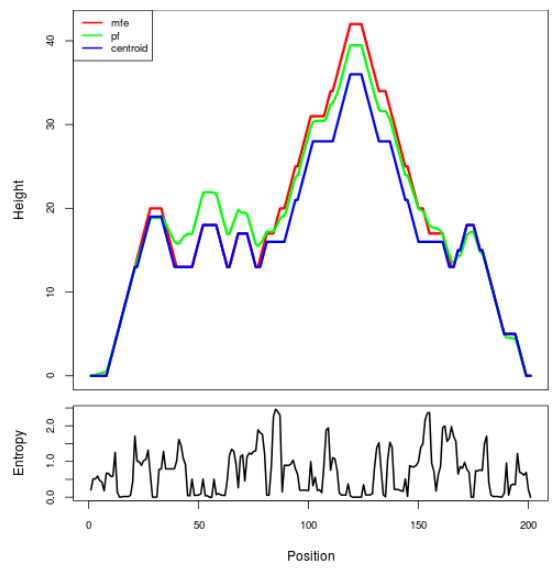
The plot represents the thermodynamic ensemble of the RNA structure of the Corynebacterium diphtheriae NCTC 13129, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
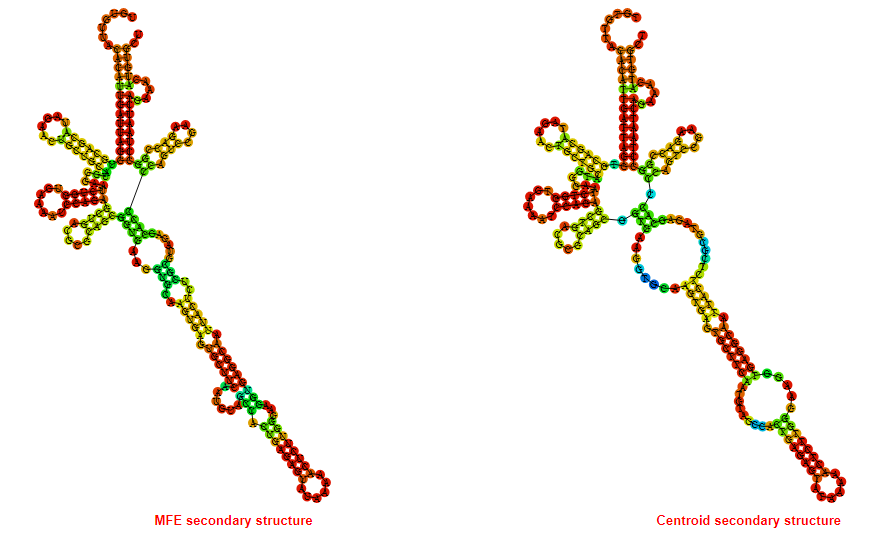
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Corynebacterium diphtheriae NCTC 13129, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
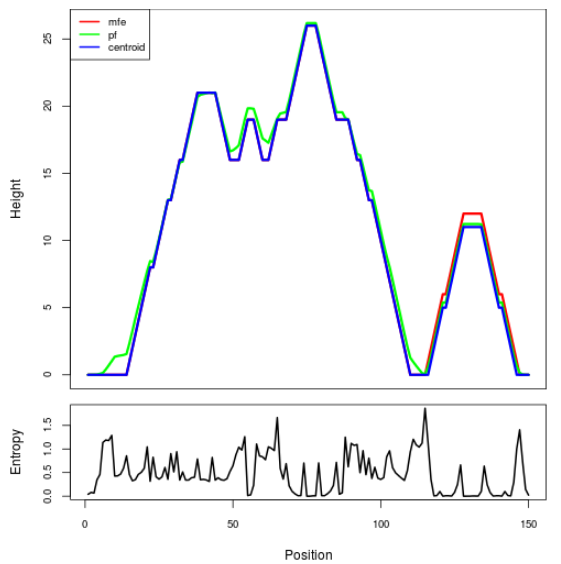
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis V583 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
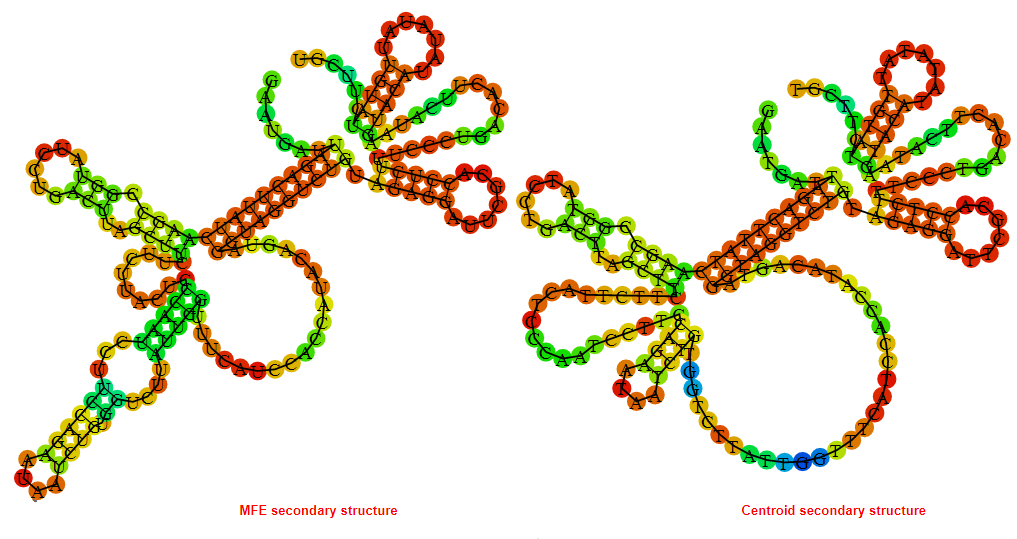
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis V583 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
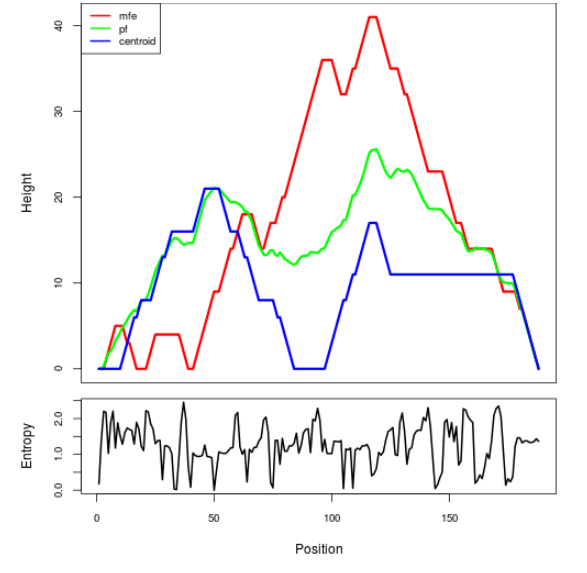
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
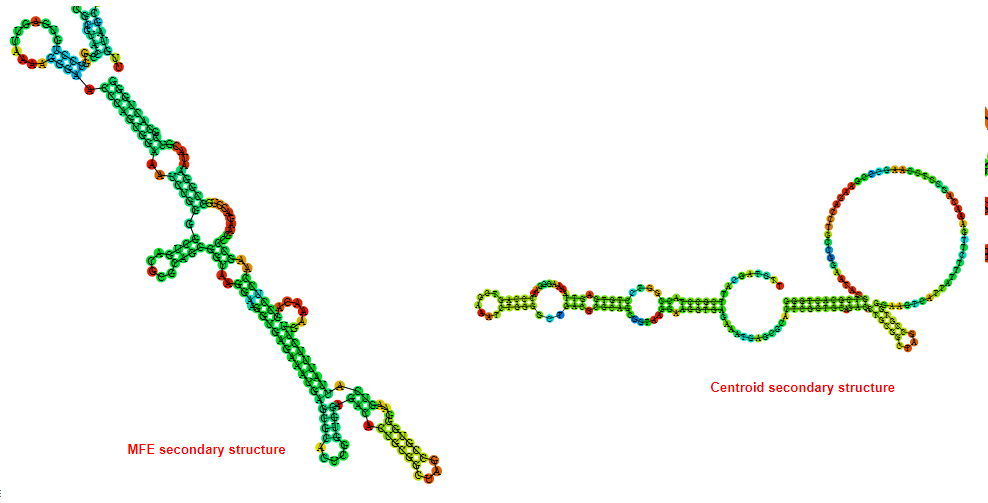
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
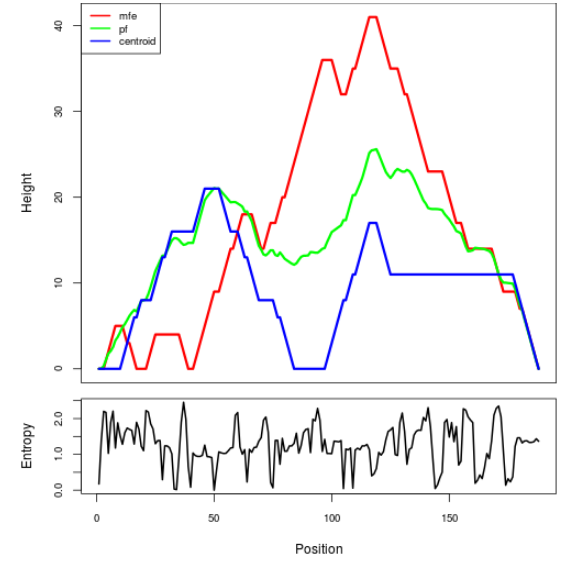
The plot represents the thermodynamic ensemble of the RNA structure of the Leptospira interrogans serovar Lai str. 56601 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
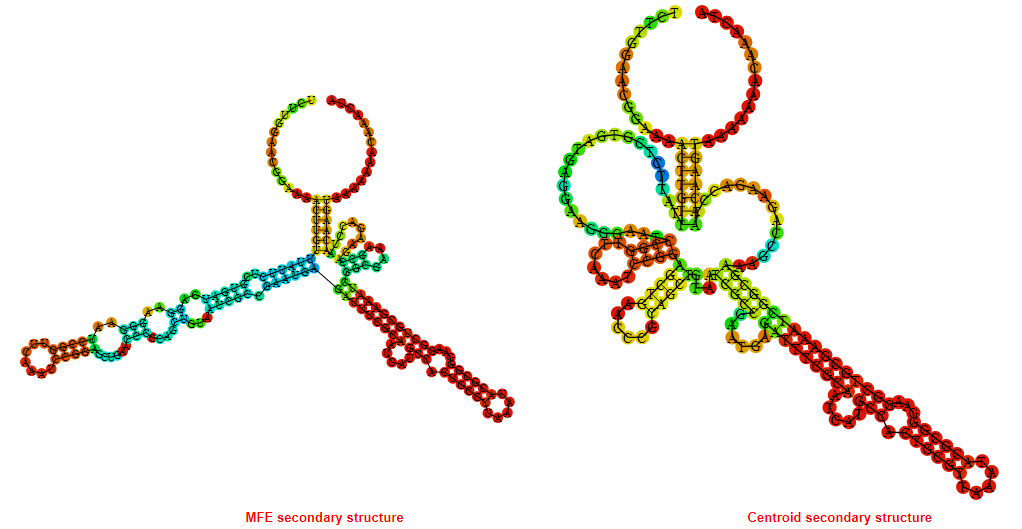
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Leptospira interrogans serovar Lai str. 56601 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
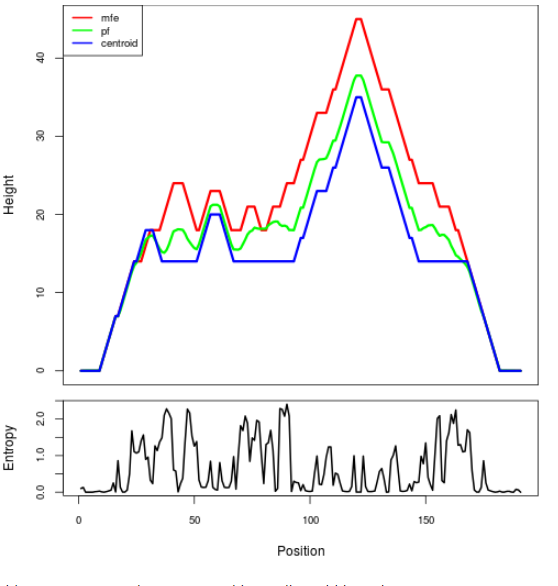
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes EGD-e, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
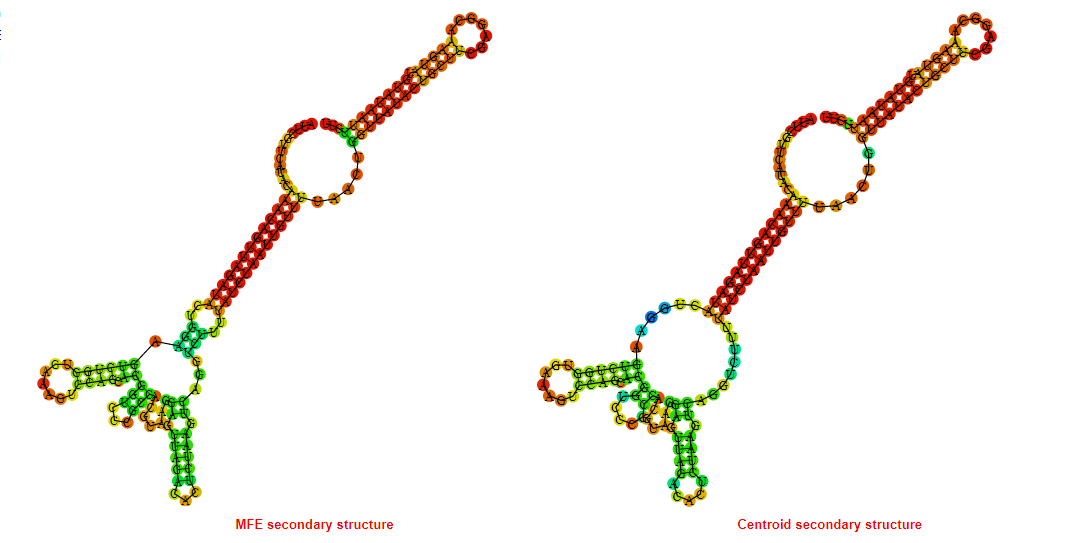
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes EGD-e, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
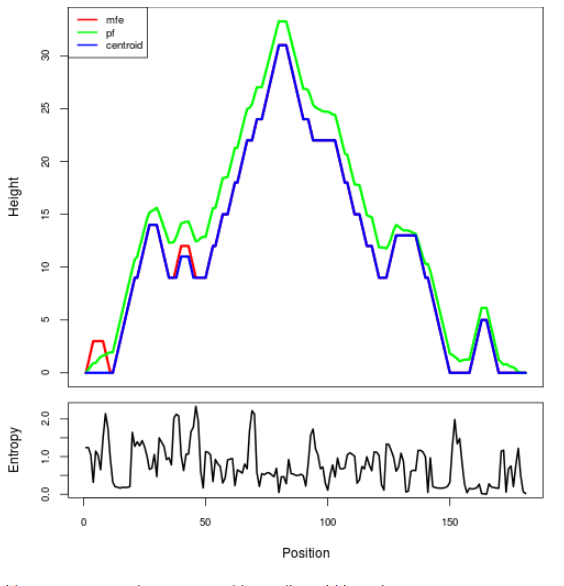
The plot represents the thermodynamic ensemble of the RNA structure of the Mycobacterium leprae TN, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
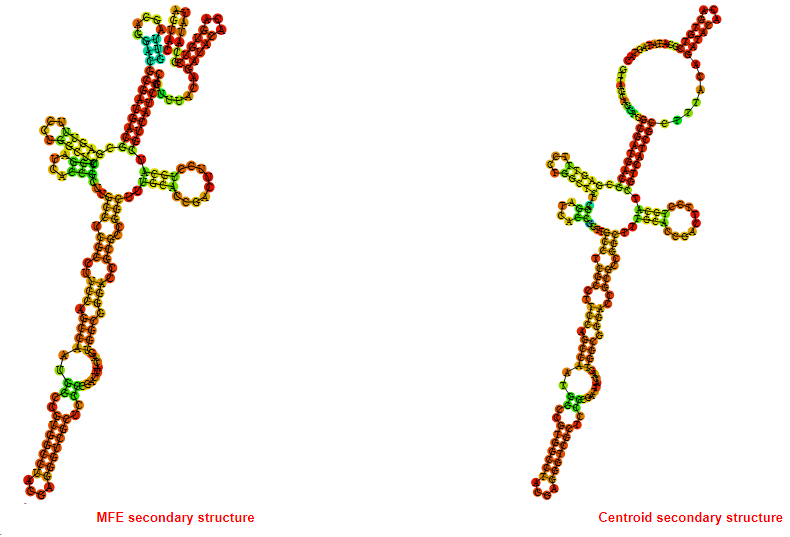
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Mycobacterium leprae TN, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
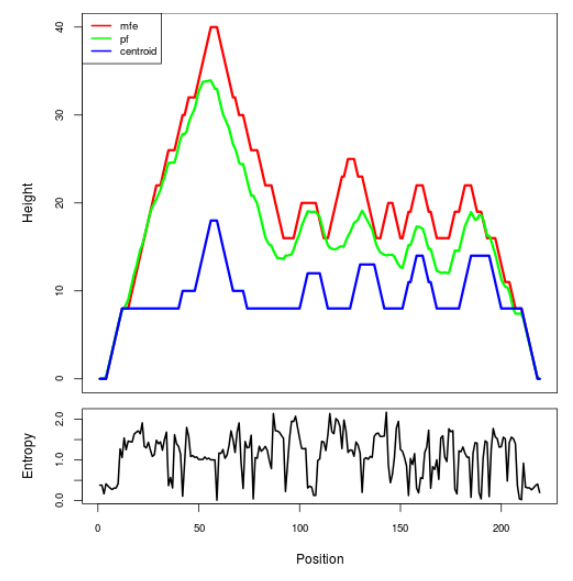
The plot represents the thermodynamic ensemble of the RNA structure of the Mycobacterium tuberculosis H37Rv, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
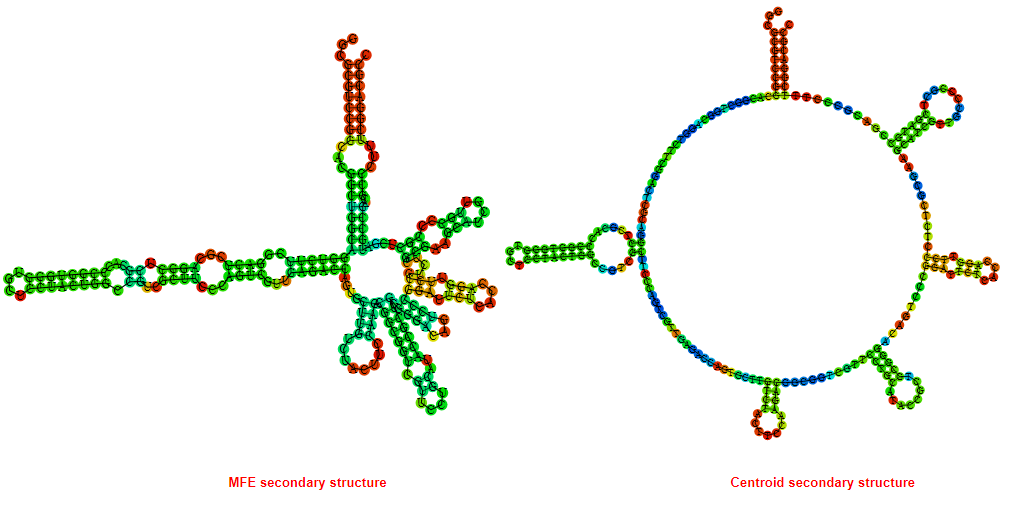
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Mycobacterium tuberculosis H37Rv, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
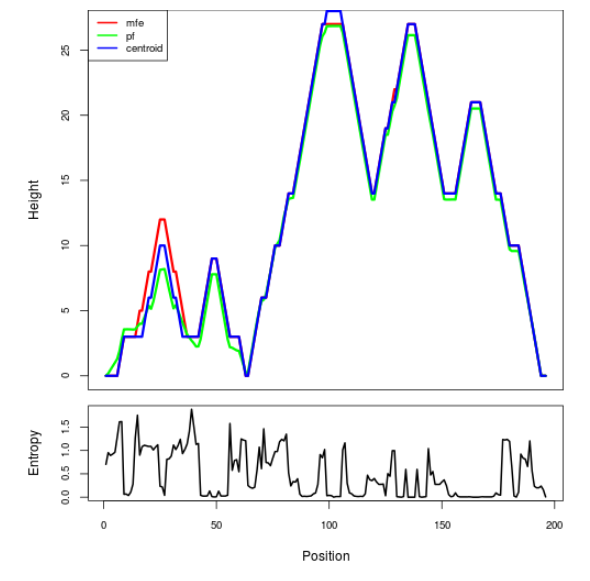
The plot represents the thermodynamic ensemble of the RNA structure of the Pseudomonas resinovorans NBRC 106553 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
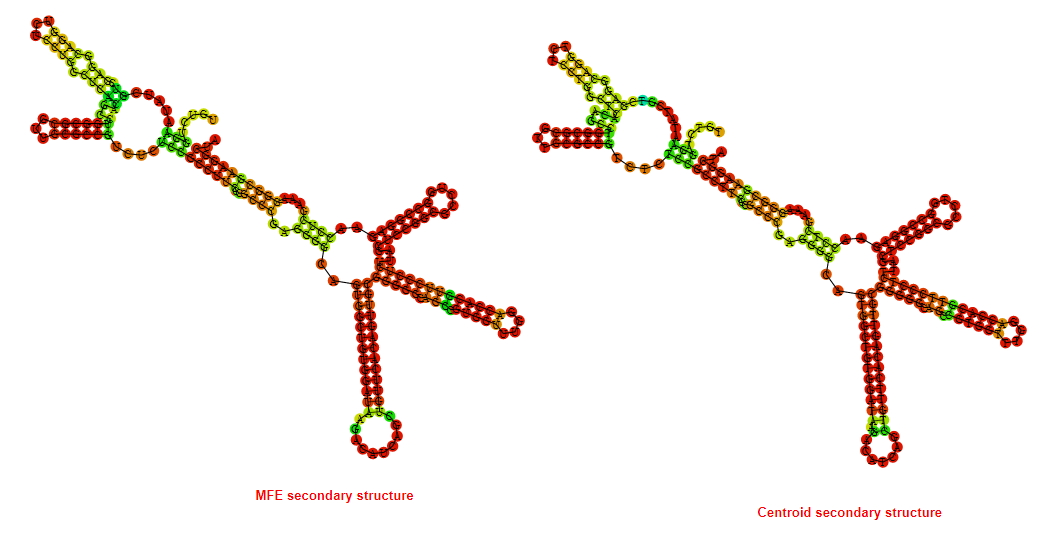
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Pseudomonas resinovorans NBRC 106553 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
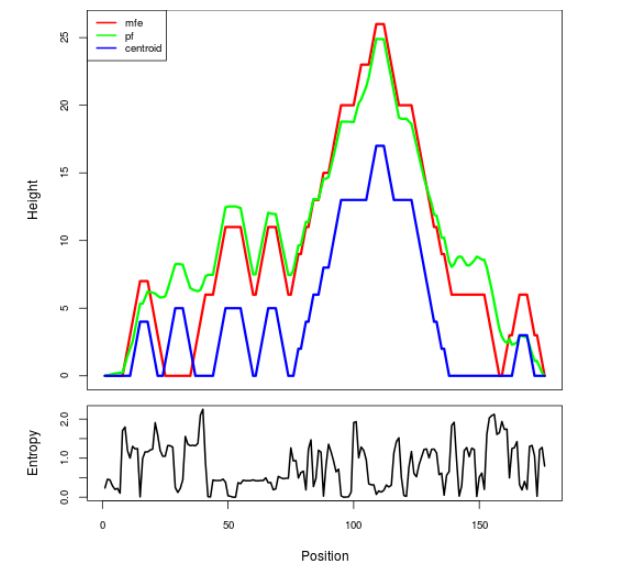
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella enterica subsp. arizonae serovar 62, Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
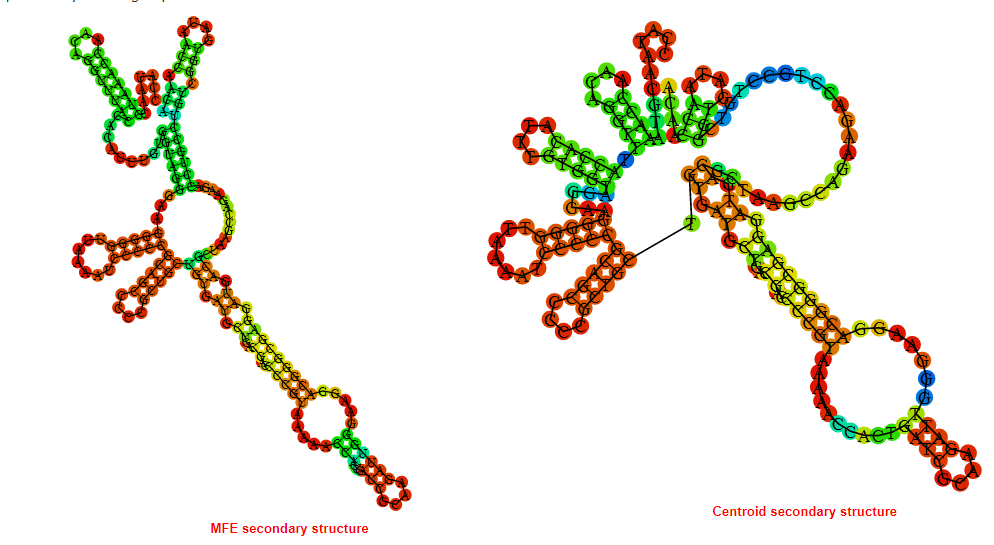
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella enterica subsp. arizonae serovar 62, Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
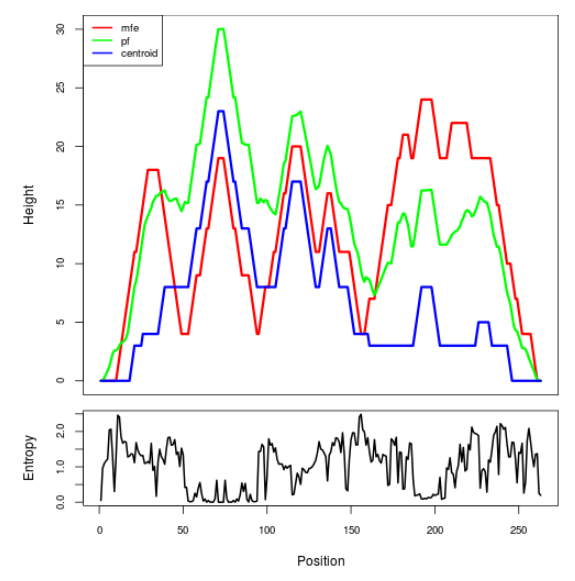
The plot represents the thermodynamic ensemble of the RNA structure of the Yersinia pestis CO92 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
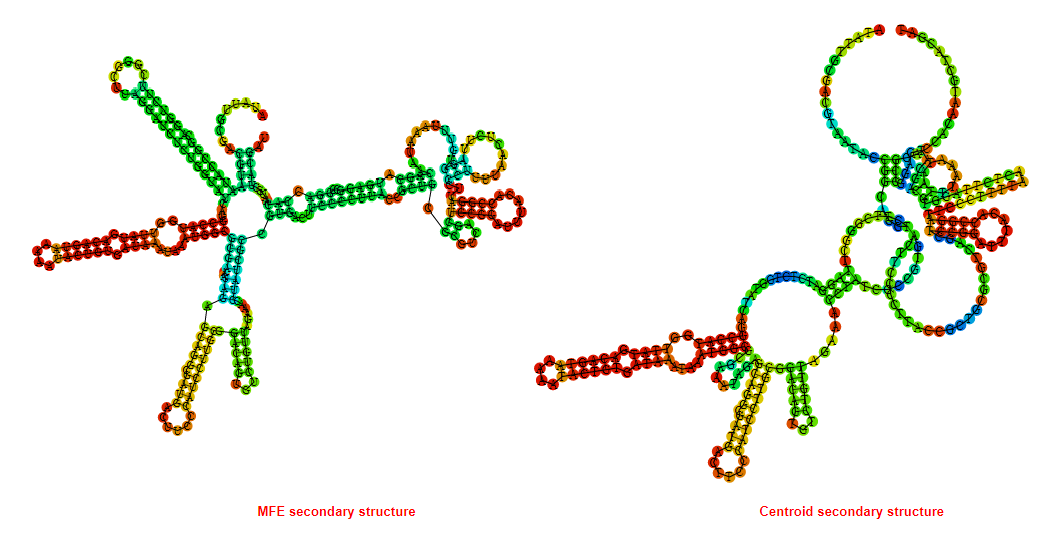
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Yersinia pestis CO92 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
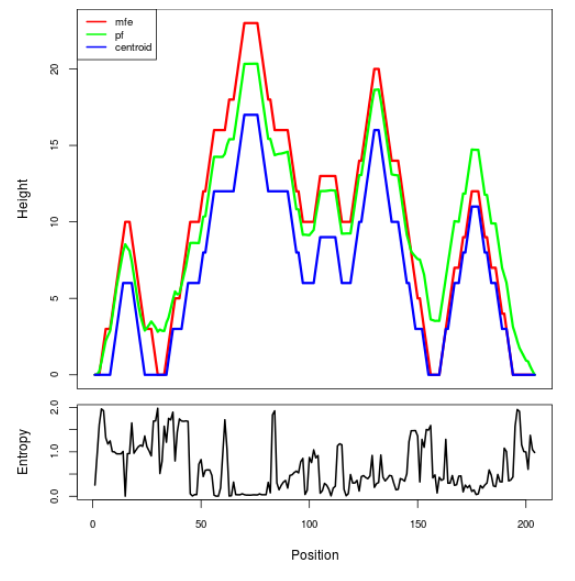
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae O1 biovar eltor str. N16961 , Cobalamin type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
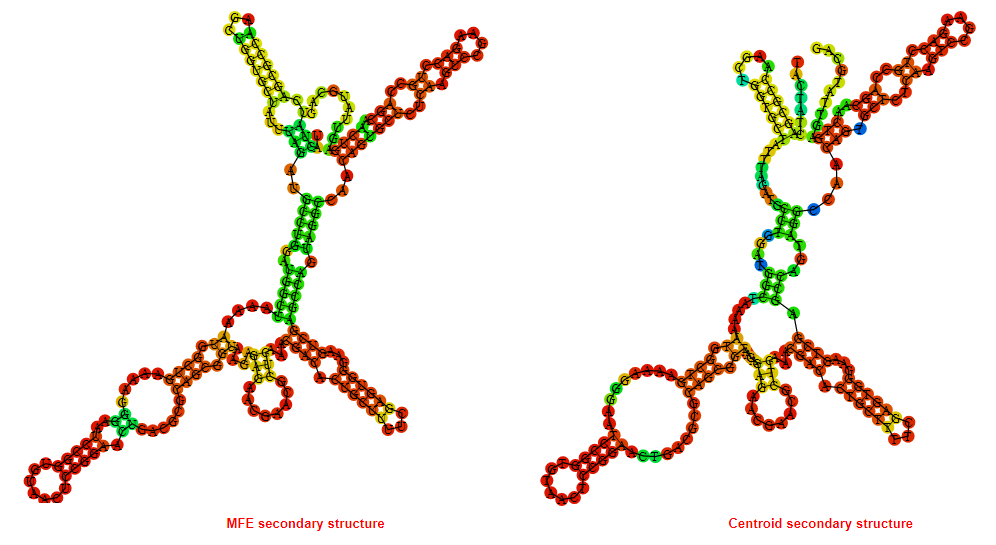
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae O1 biovar eltor str. N16961 , Cobalamin type switch.
Multiple alignment of the Cobalamin type switches
Below is presented the sequences alignment of all the Cobalamin type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cobalamin type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cobalamin type switches involved in.

Multi_resistance strains for the Cobalamin type switches
The table displays known human pathogenic bacteria bearing the Cobalamin type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
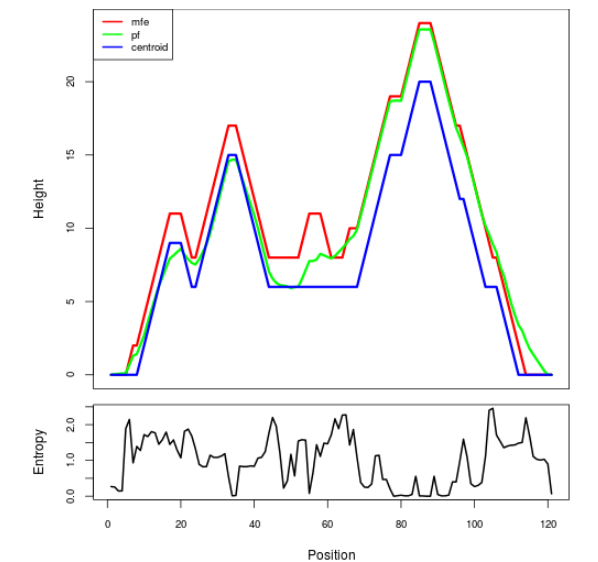
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis, Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
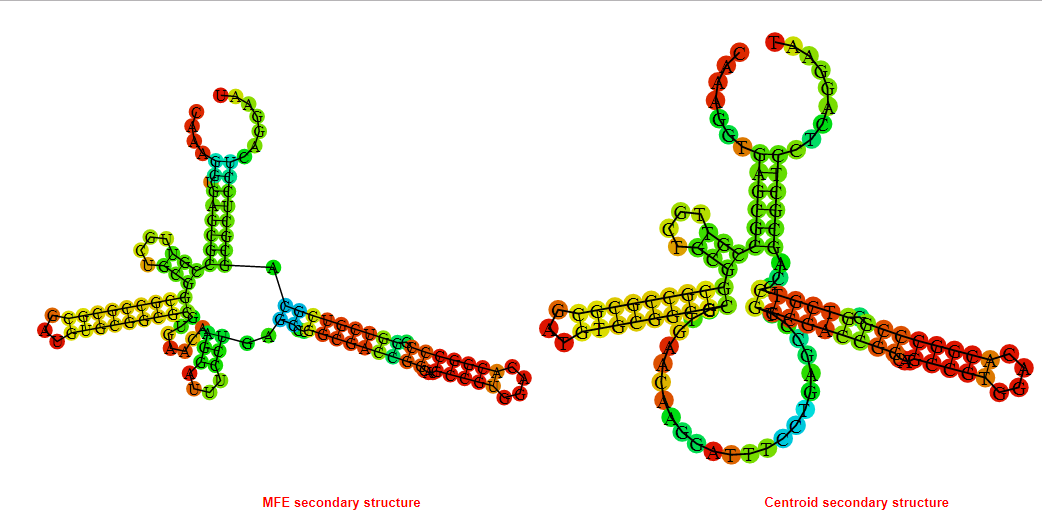
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis, Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.
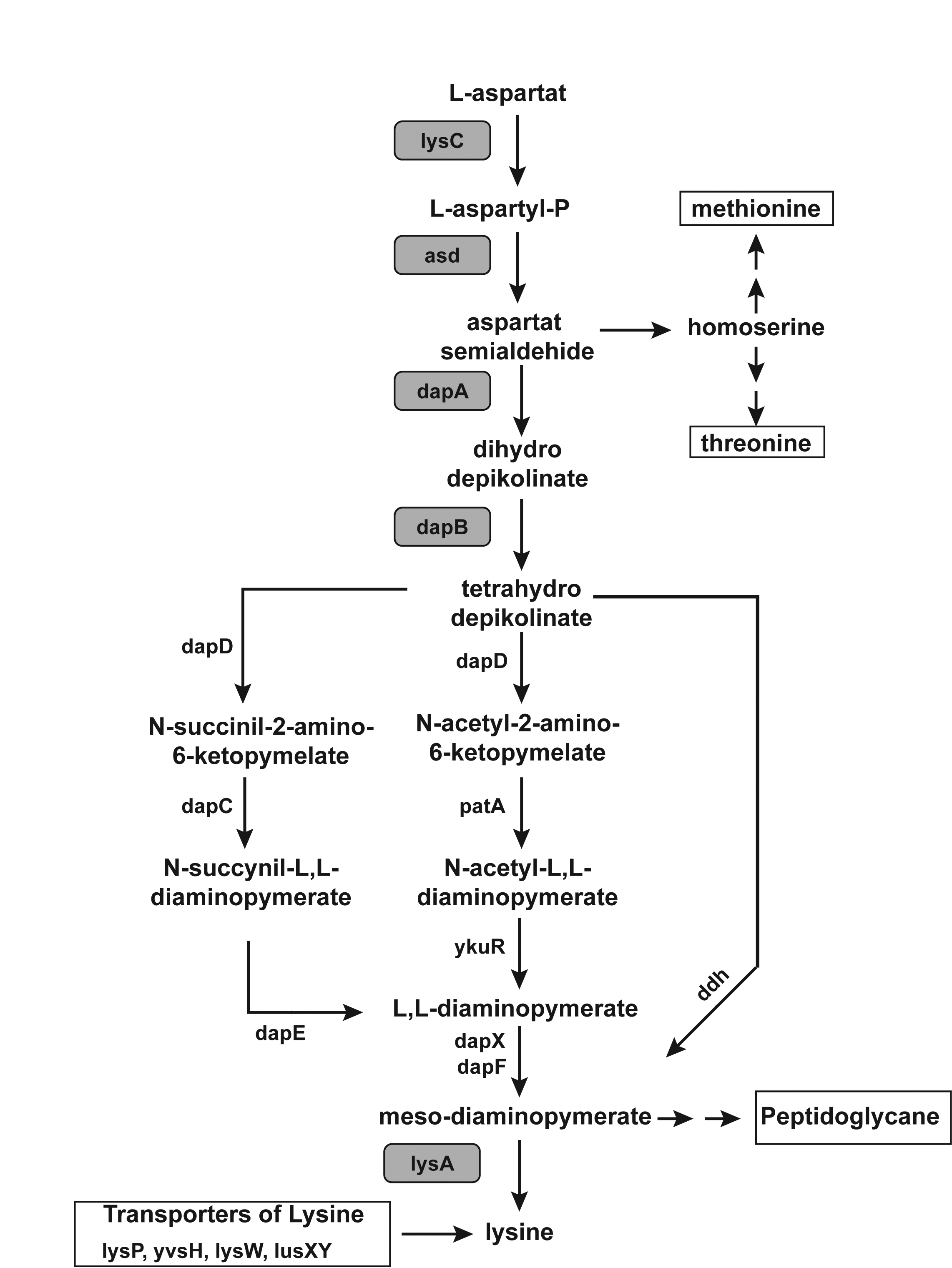
Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
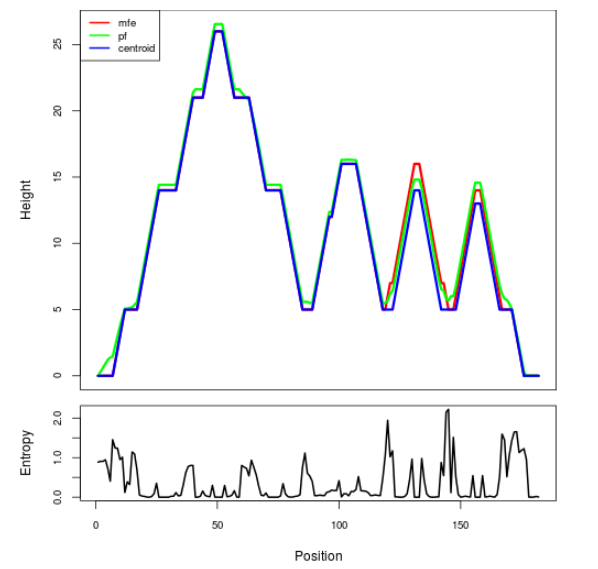
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus cereus, Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
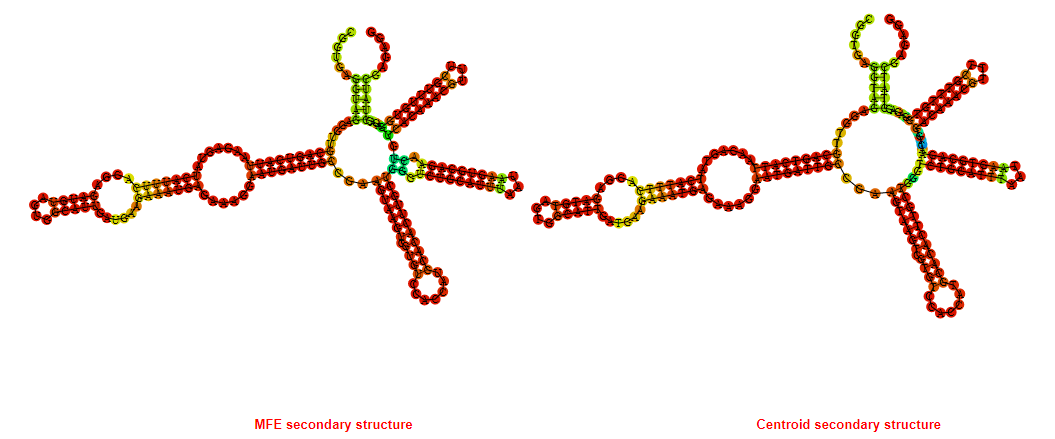
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus cereus, Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
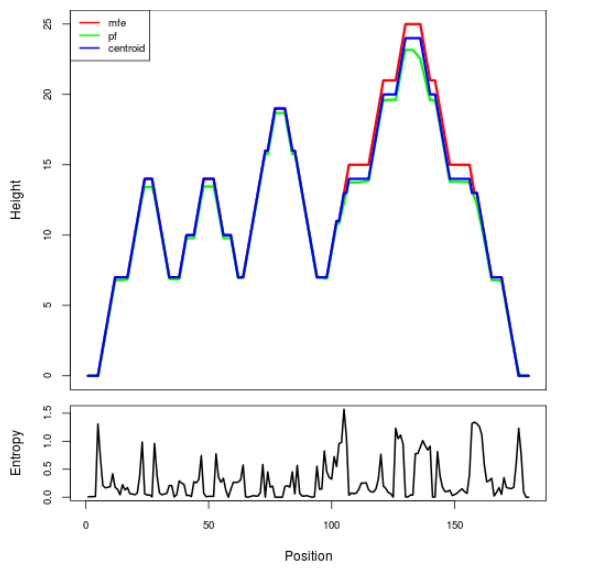
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
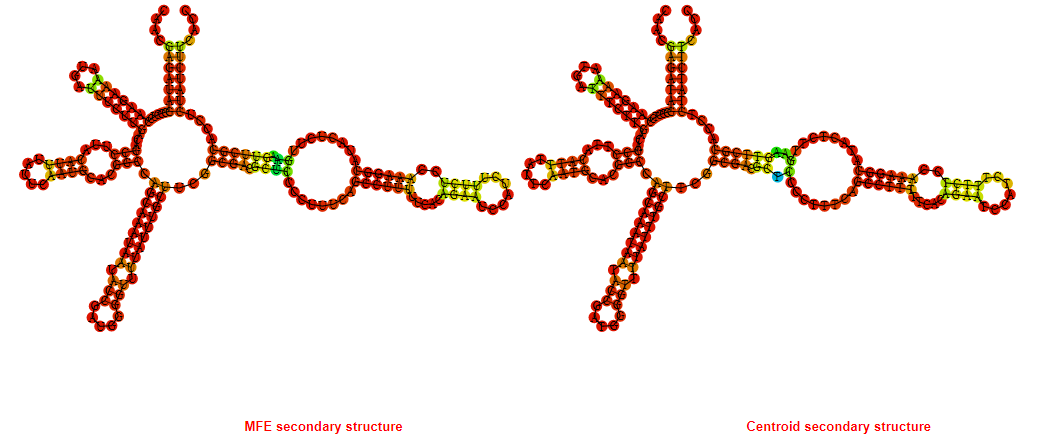
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
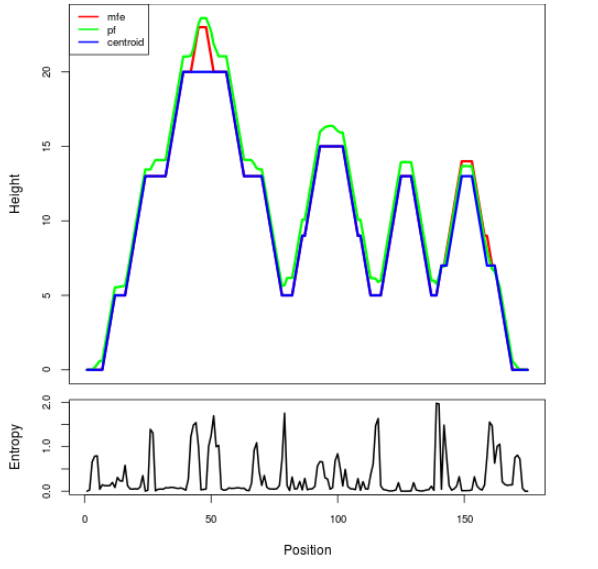
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
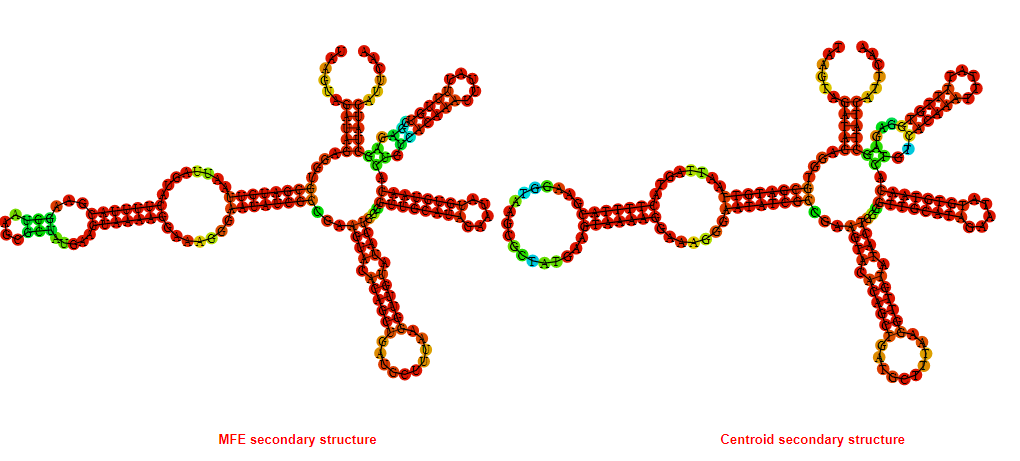
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
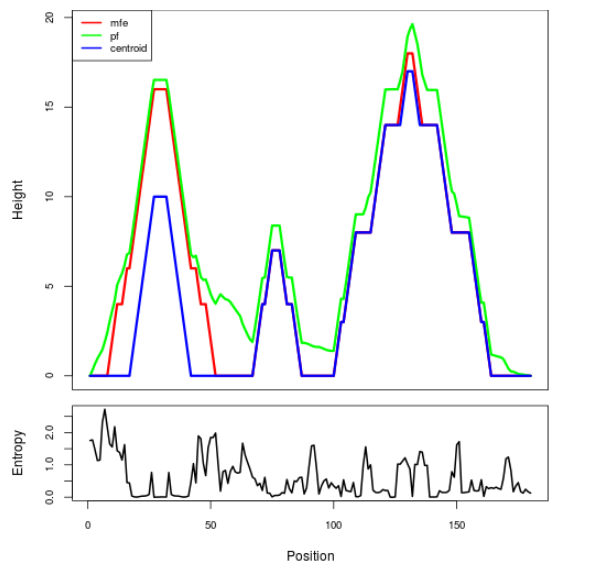
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
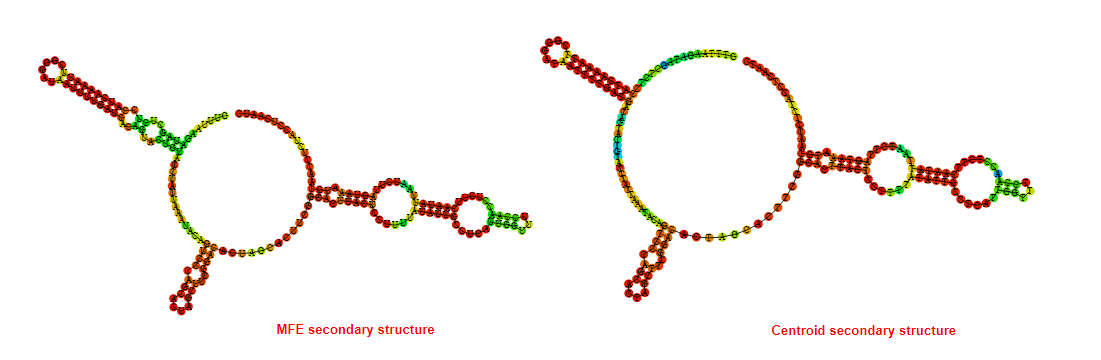
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
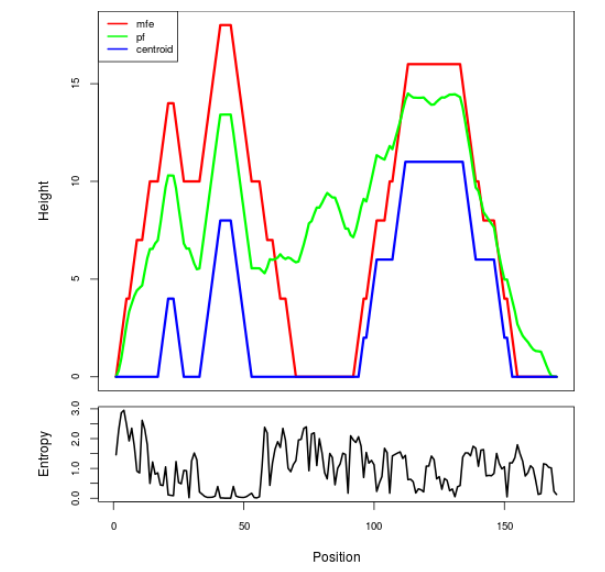
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
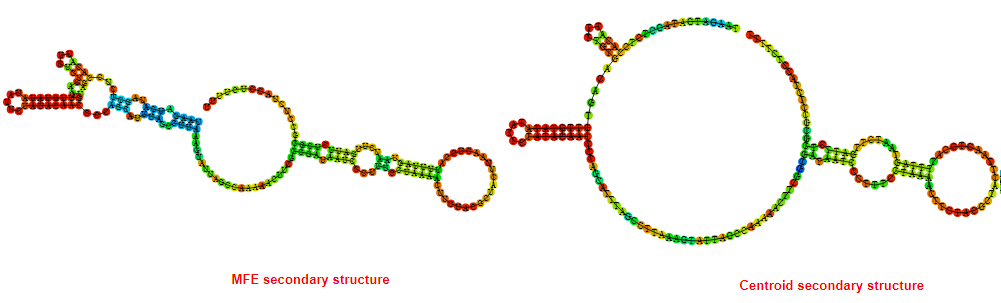
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
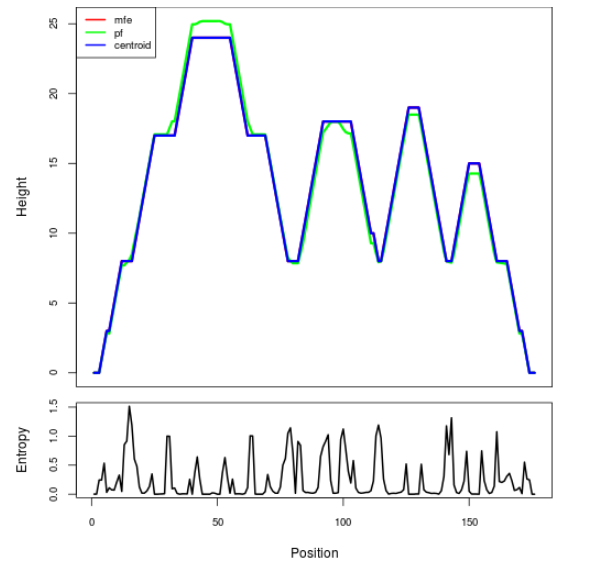
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
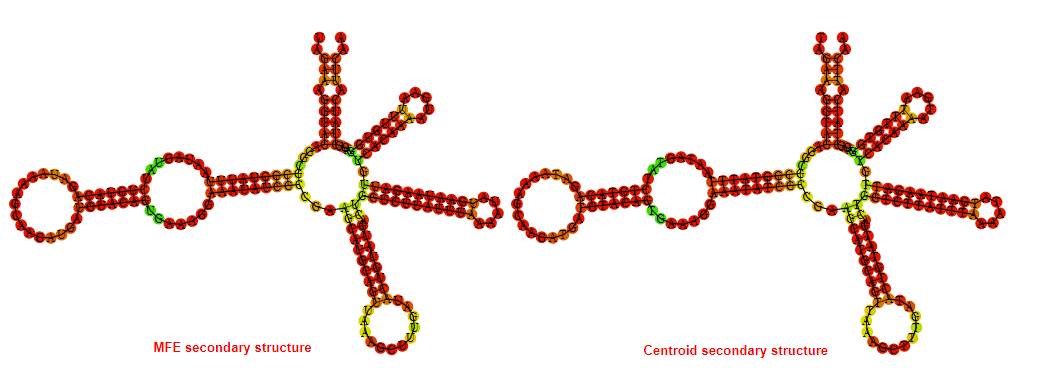
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
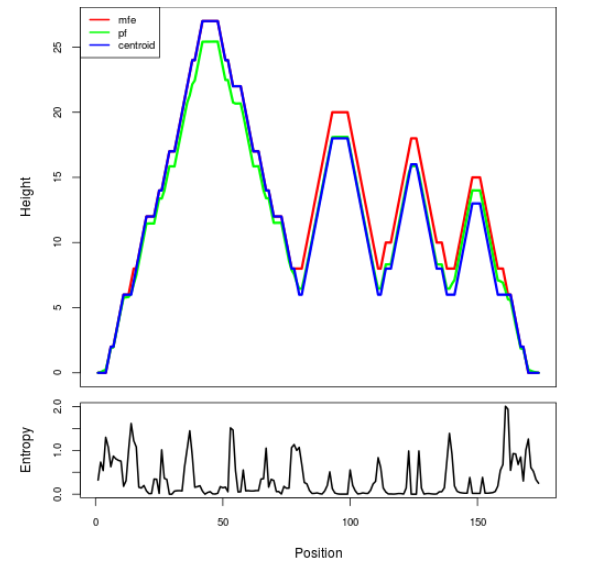
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
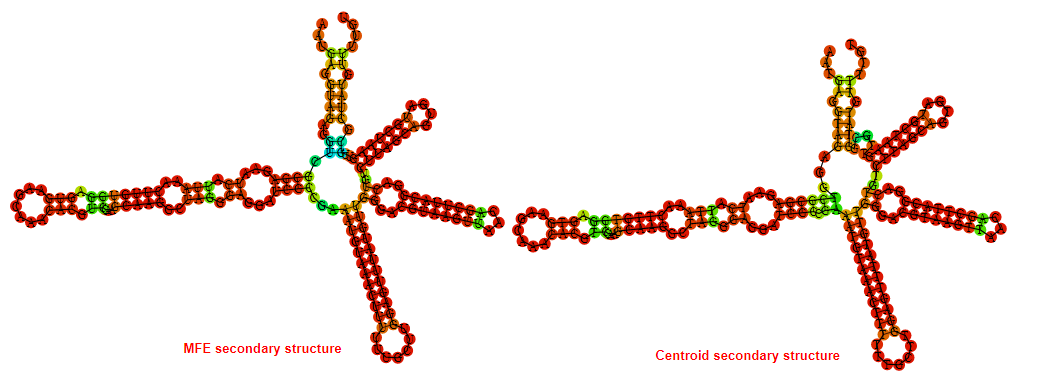
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
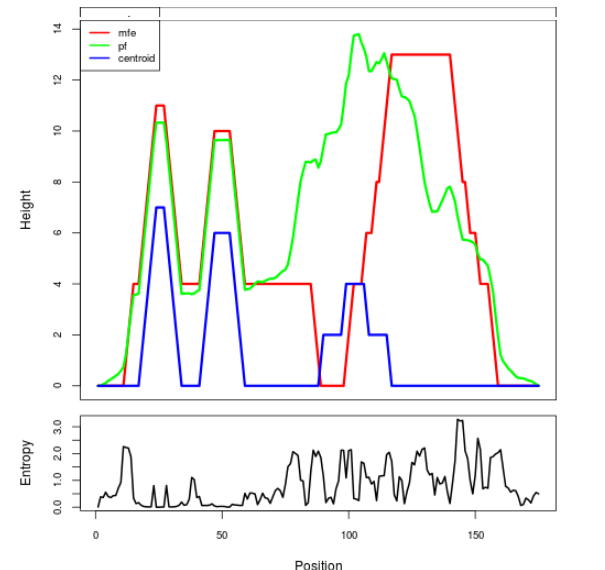
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
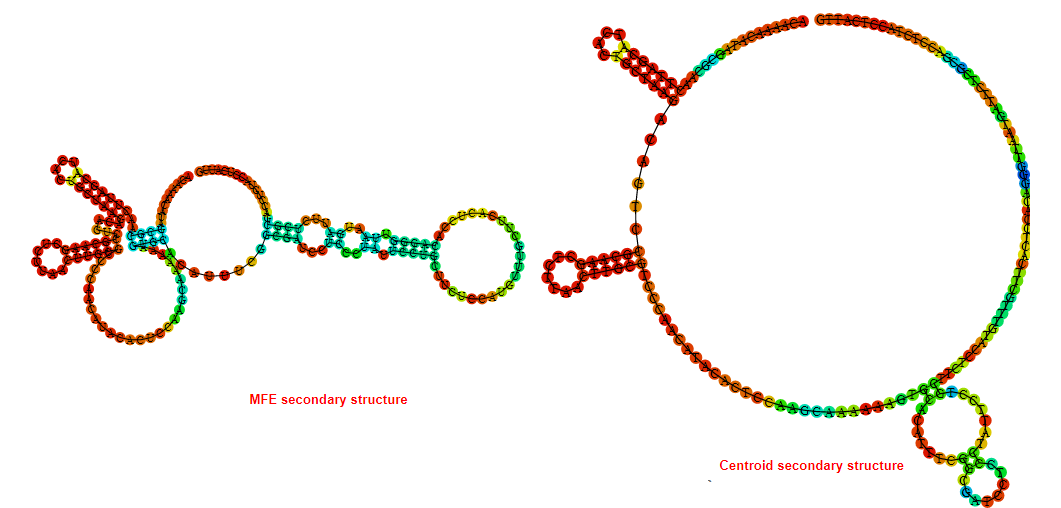
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
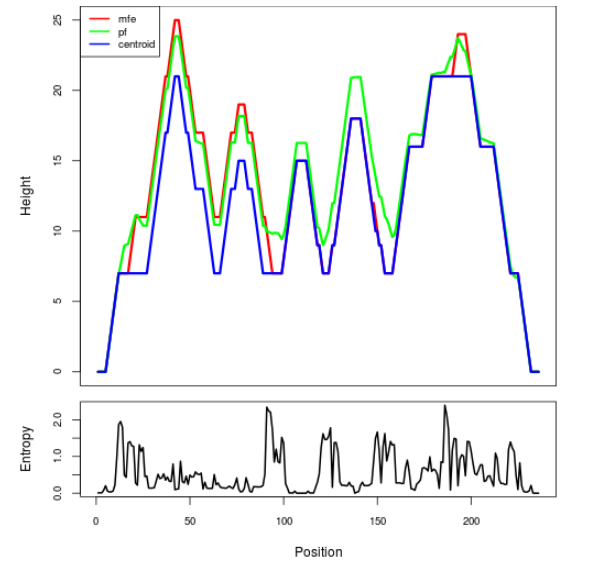
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
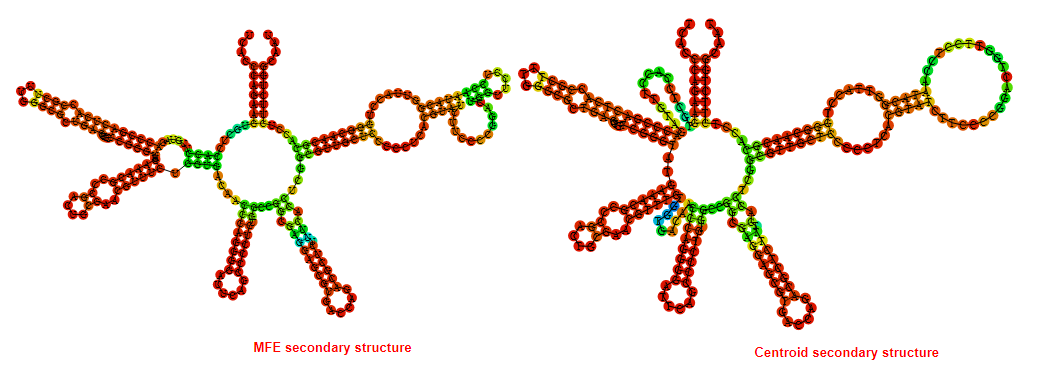
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
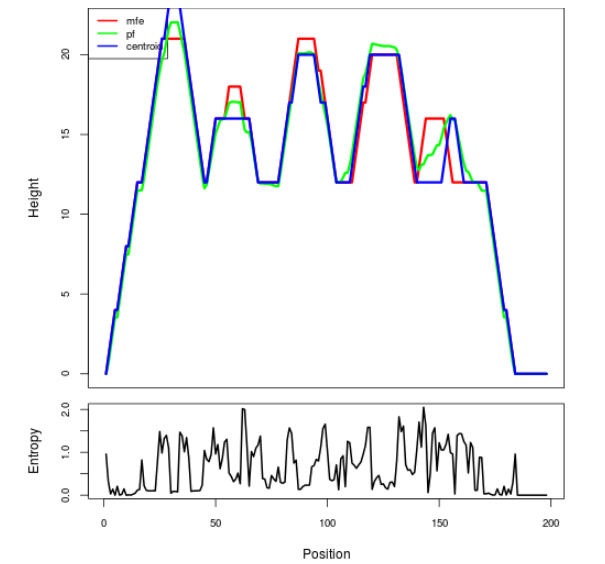
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
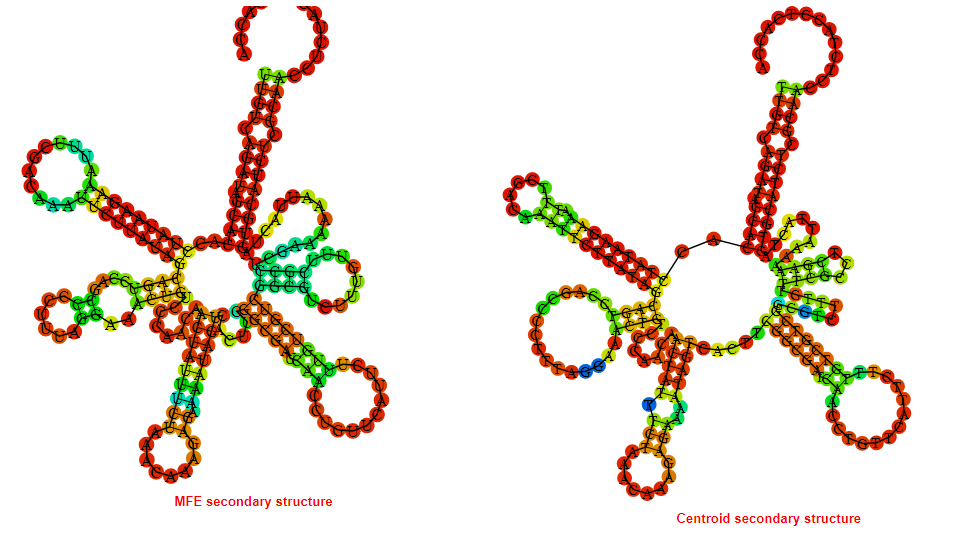
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
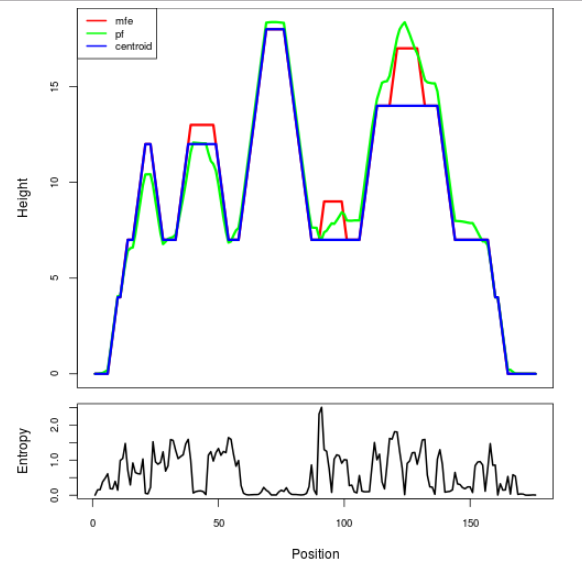
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
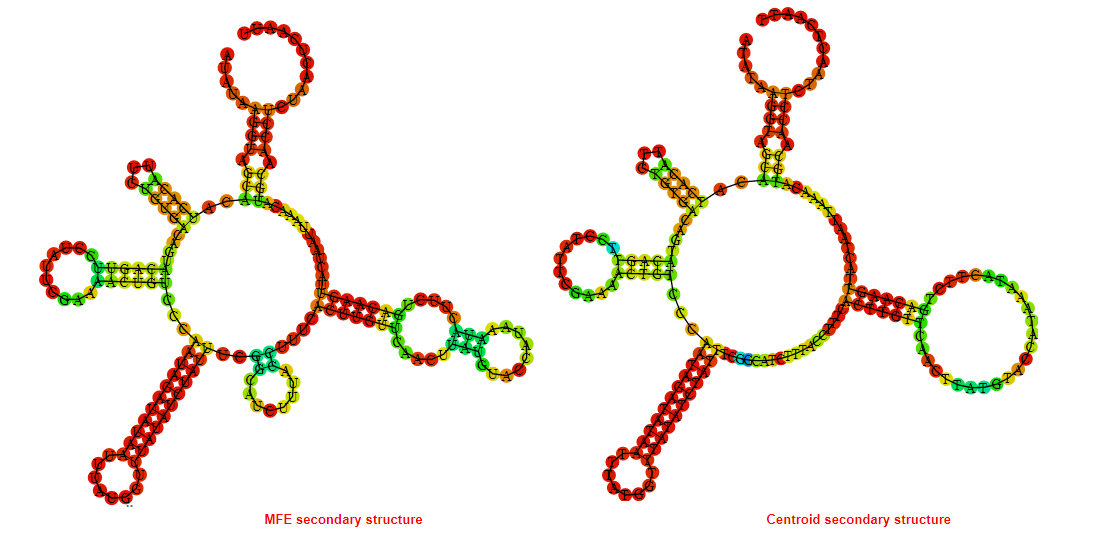
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
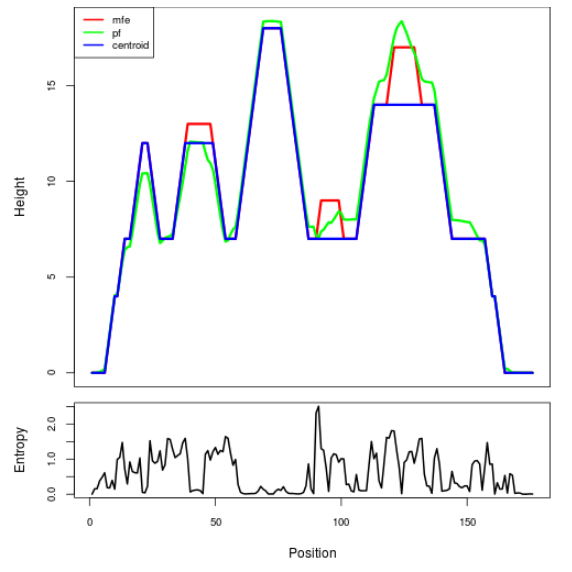
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
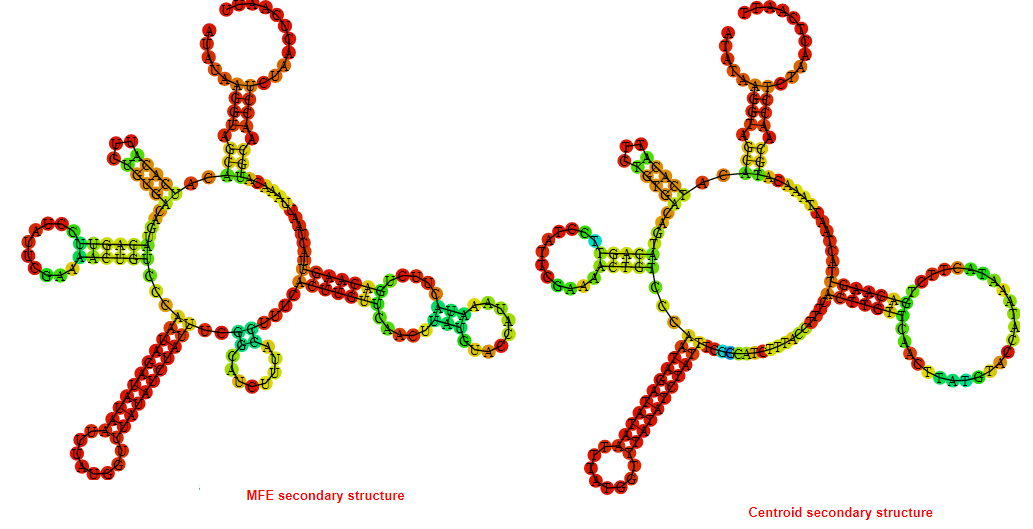
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
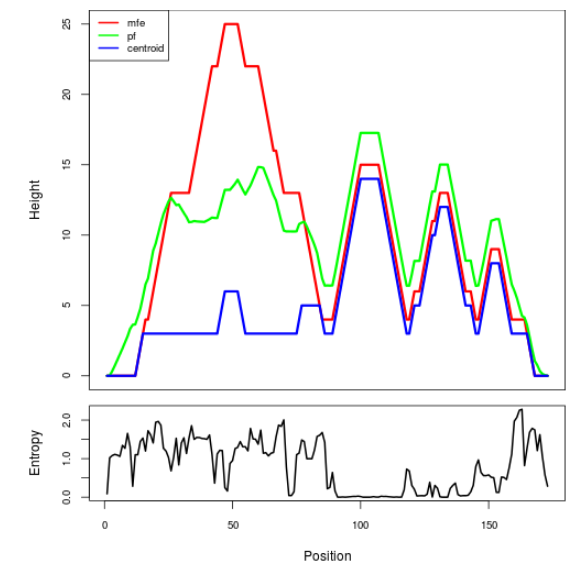
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
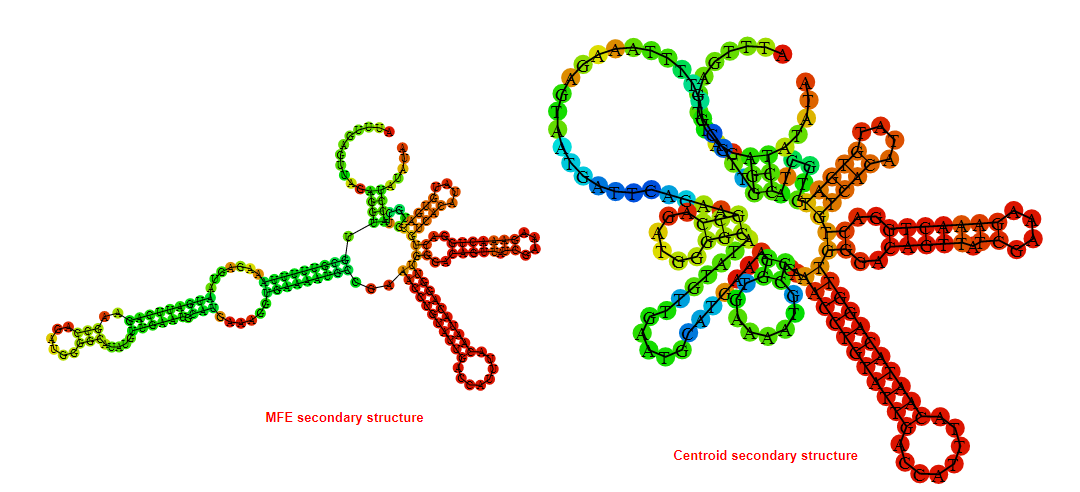
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
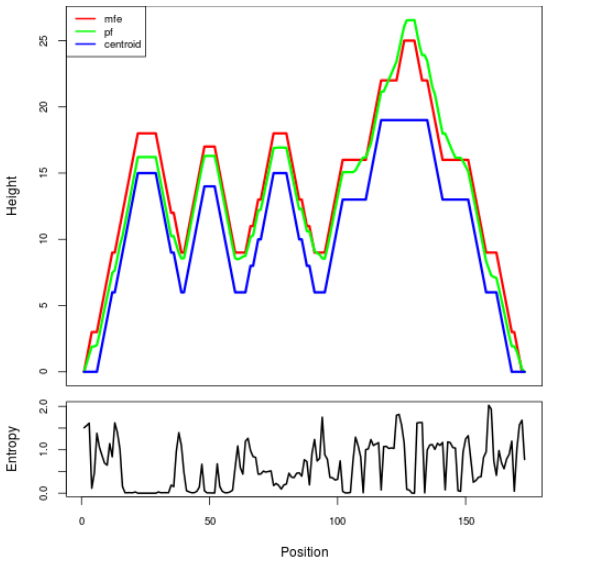
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
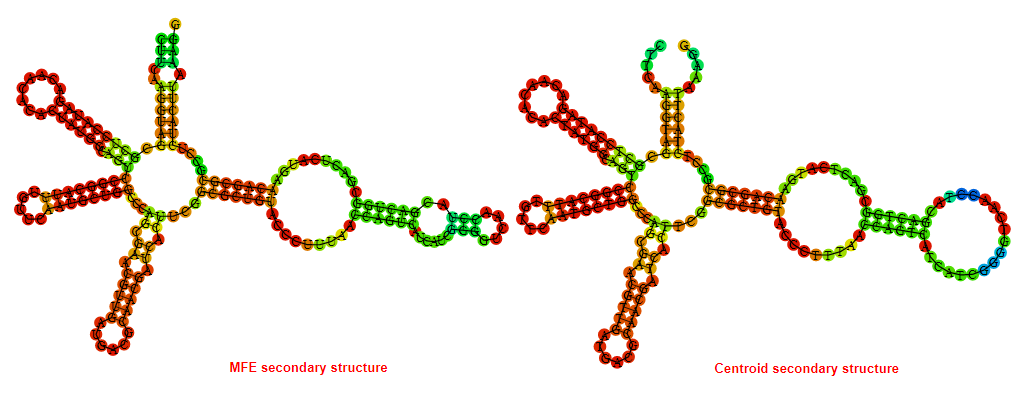
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
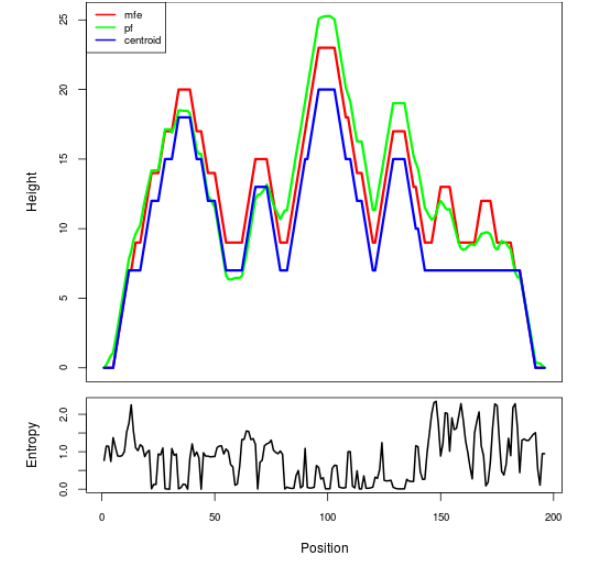
The plot represents the thermodynamic ensemble of the RNA structure of the Escherichia coli , Lysine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
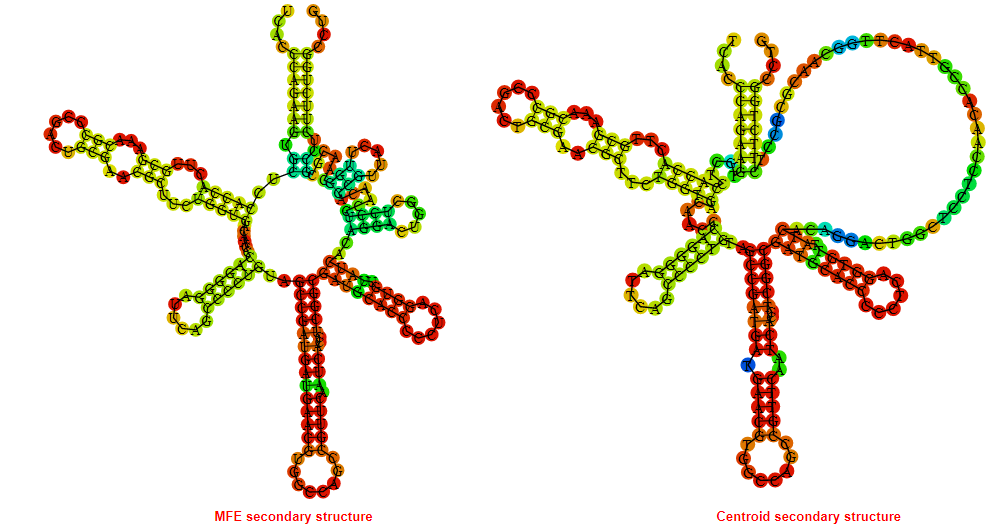
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Escherichia coli , Lysine type switch.
Multiple alignment of the Lysine type switches
Below is presented the sequences alignment of all the Lysine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Lysine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Lysine type switches involved in.

Multi_resistance strains for the Lysine type switches
The table displays known human pathogenic bacteria bearing the Lysine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
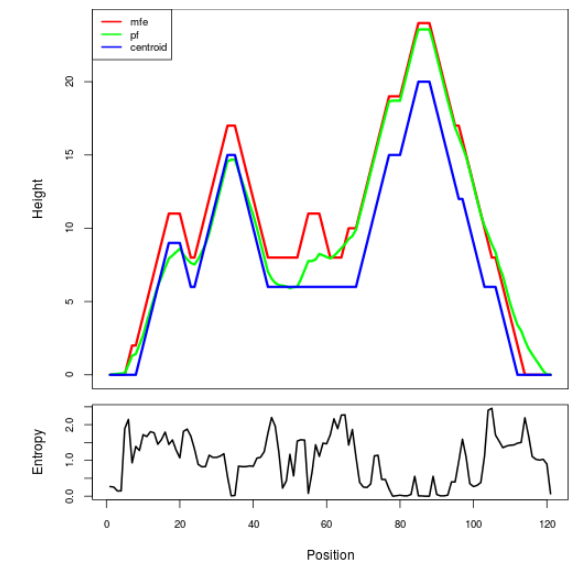
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis , SAH type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
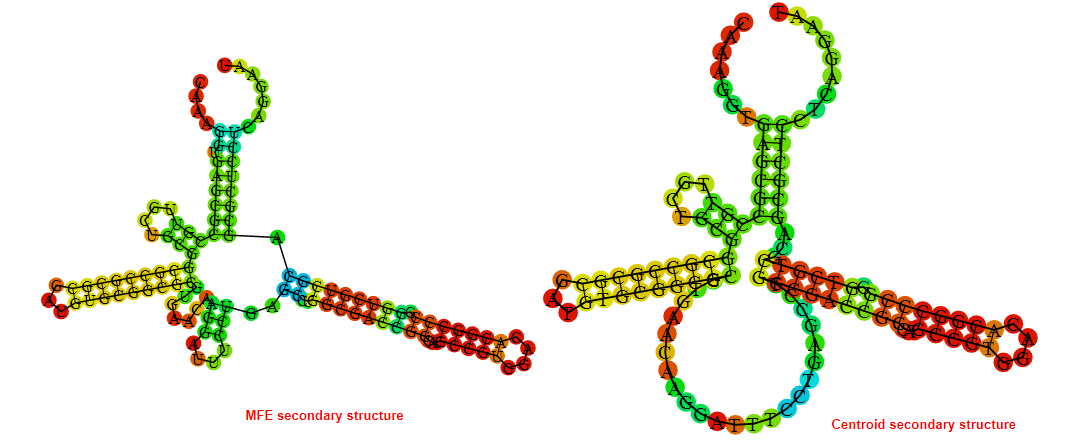
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis , SAH type switch.
Multiple alignment of the SAH type switches
Below is presented the sequences alignment of all the SAH type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAH type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAH type switches involved in.
Multi_resistance strains for the SAH type switches
The table displays known human pathogenic bacteria bearing the SAH type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
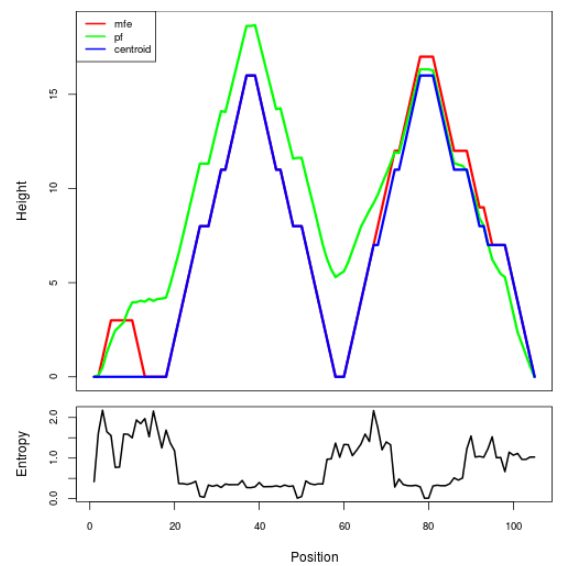
The plot represents the thermodynamic ensemble of the RNA structure of the Pseudomonas aeruginosa , SAH type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
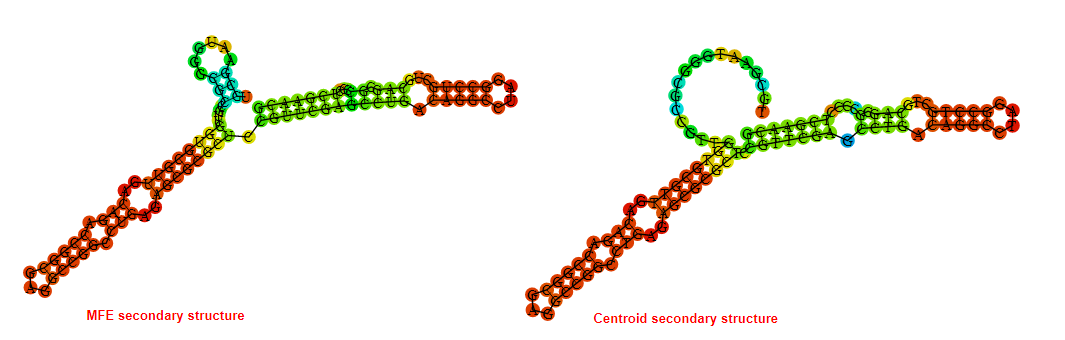
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Pseudomonas aeruginosa , SAH type switch.
Multiple alignment of the SAH type switches
Below is presented the sequences alignment of all the SAH type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAH type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAH type switches involved in.

Multi_resistance strains for the SAH type switches
The table displays known human pathogenic bacteria bearing the SAH type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
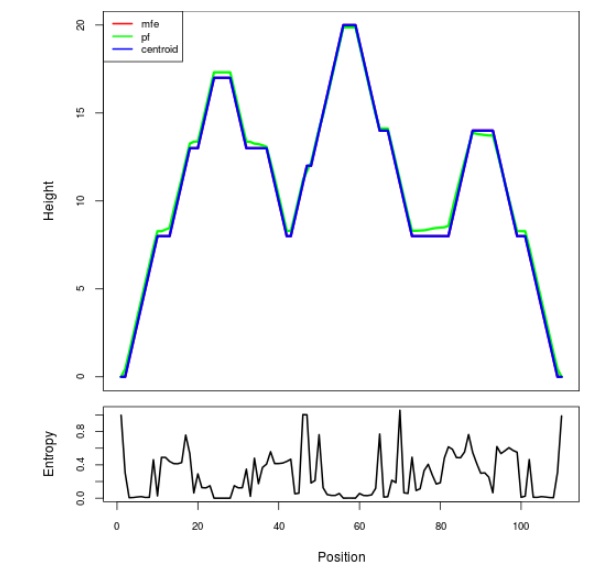
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
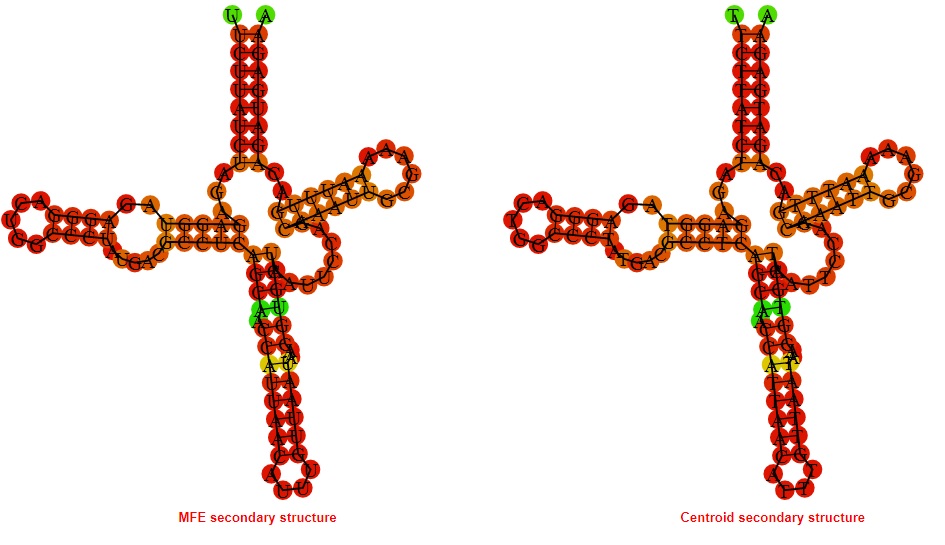
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.
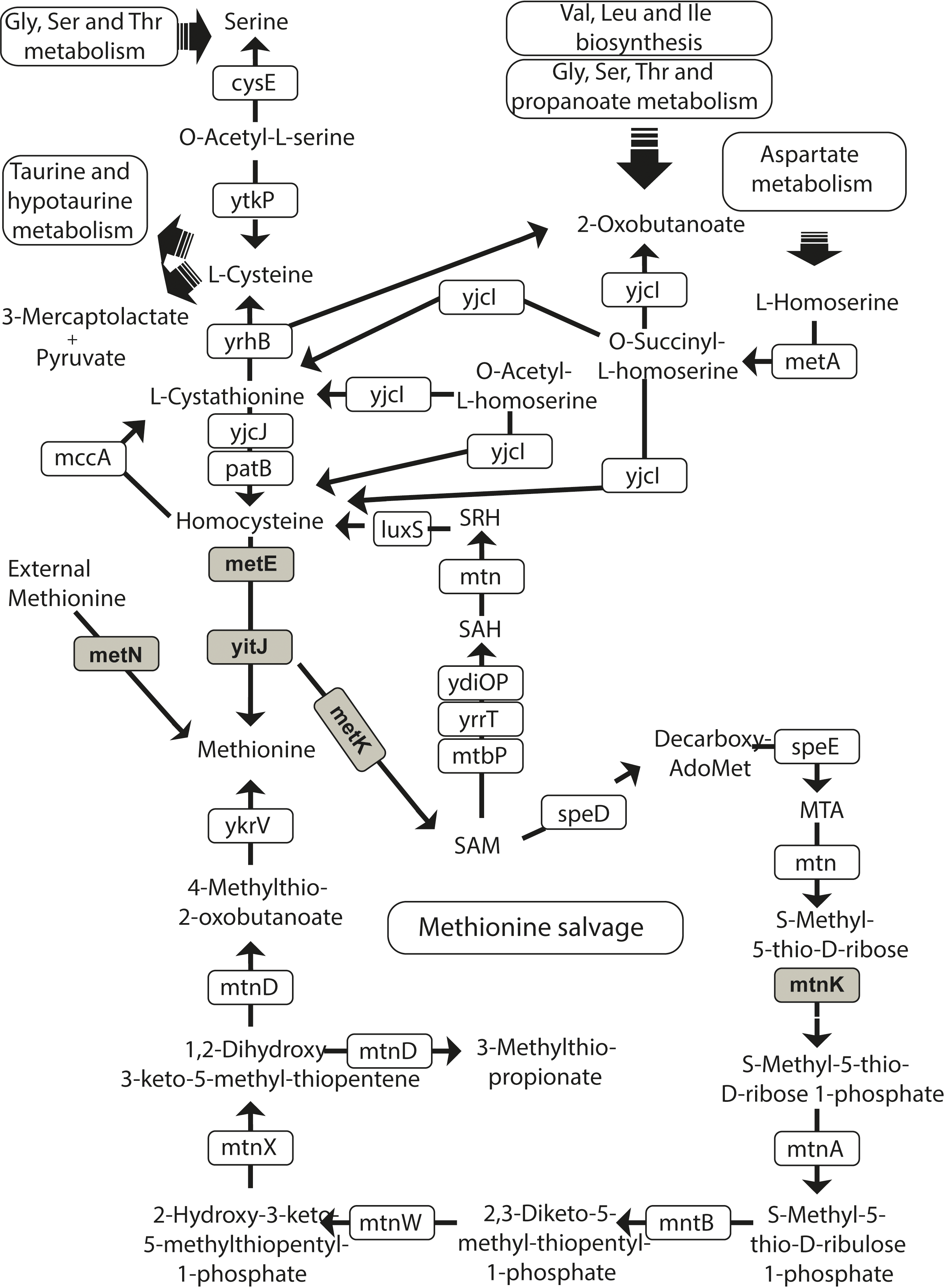
Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
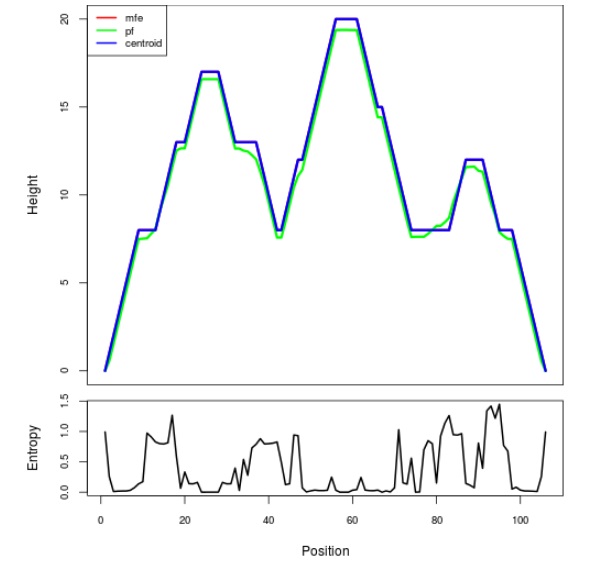
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus cereus, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
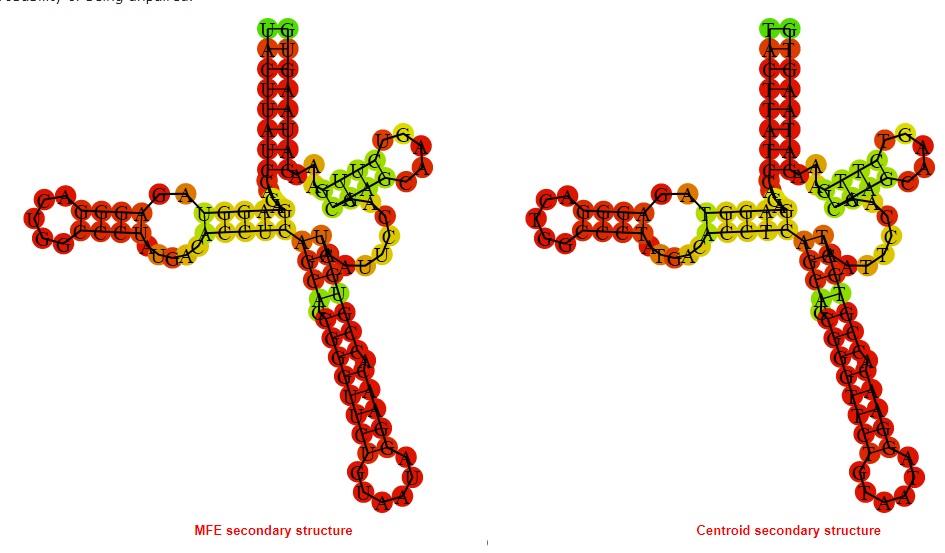
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus cereus, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
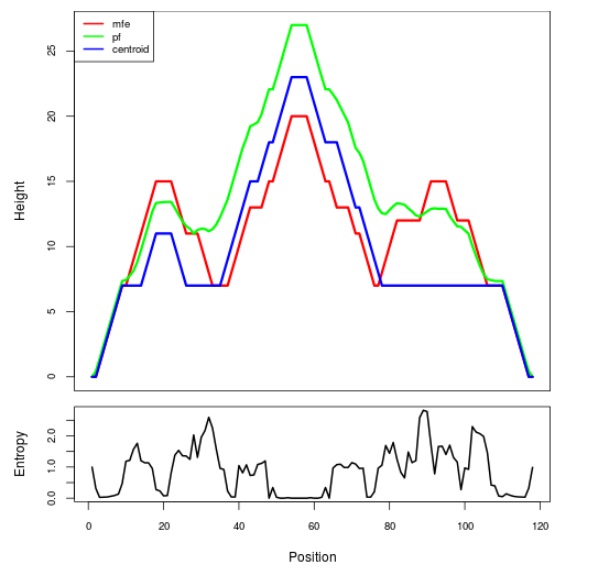
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
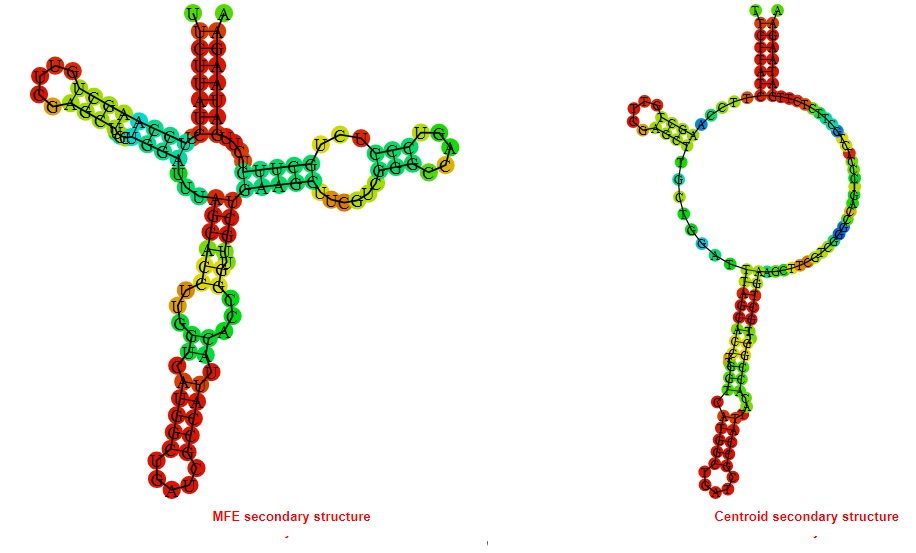
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
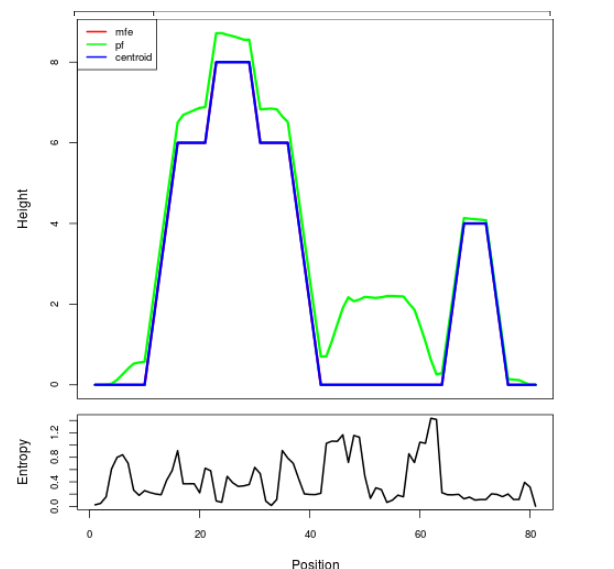
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
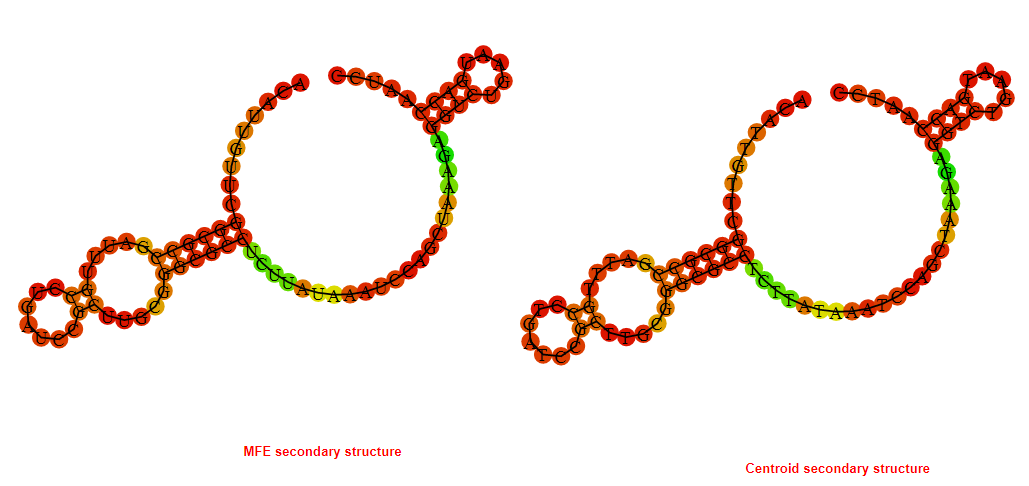
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
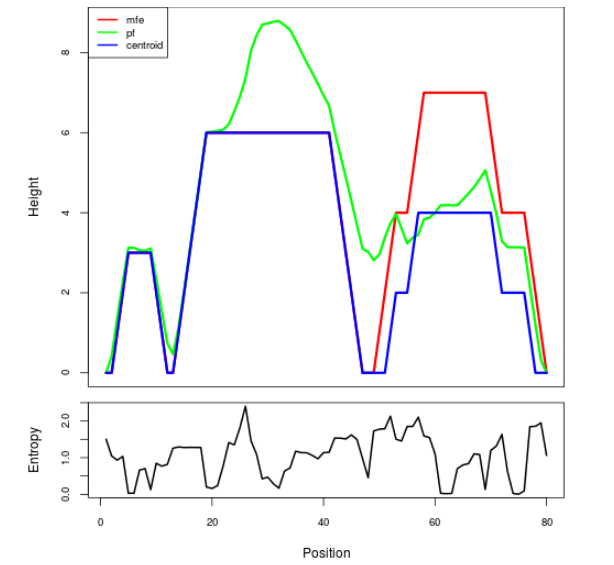
The plot represents the thermodynamic ensemble of the RNA structure of the Brucella abortis , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
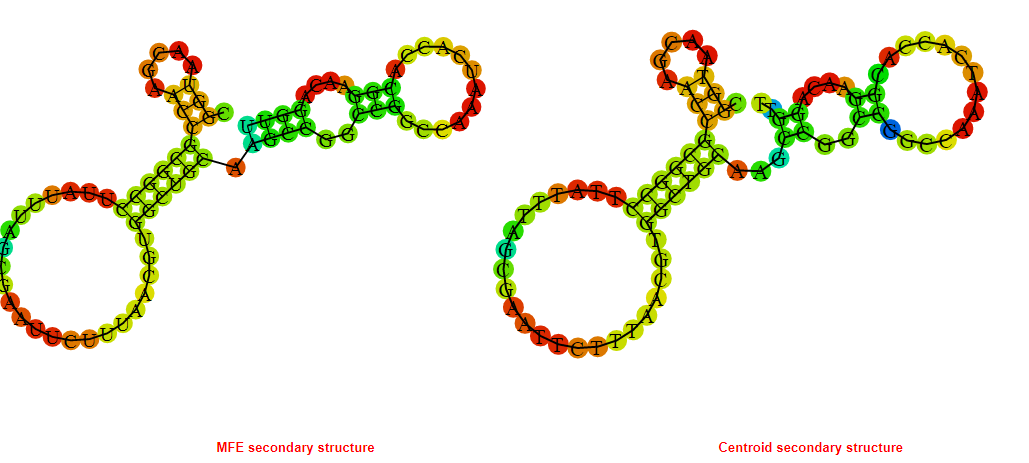
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Brucella abortis , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
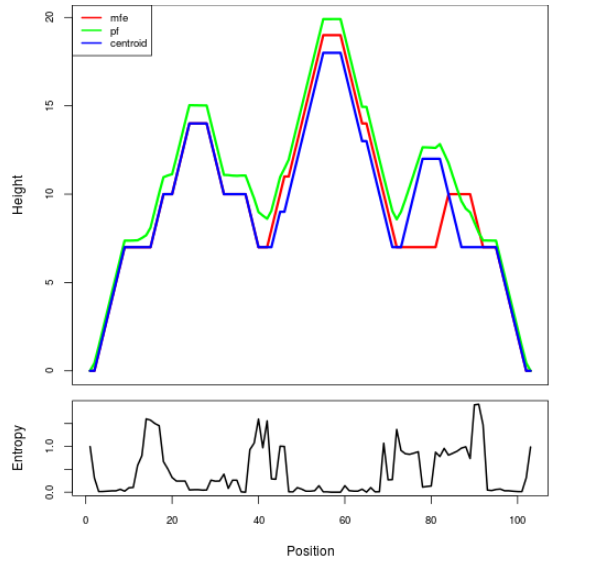
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
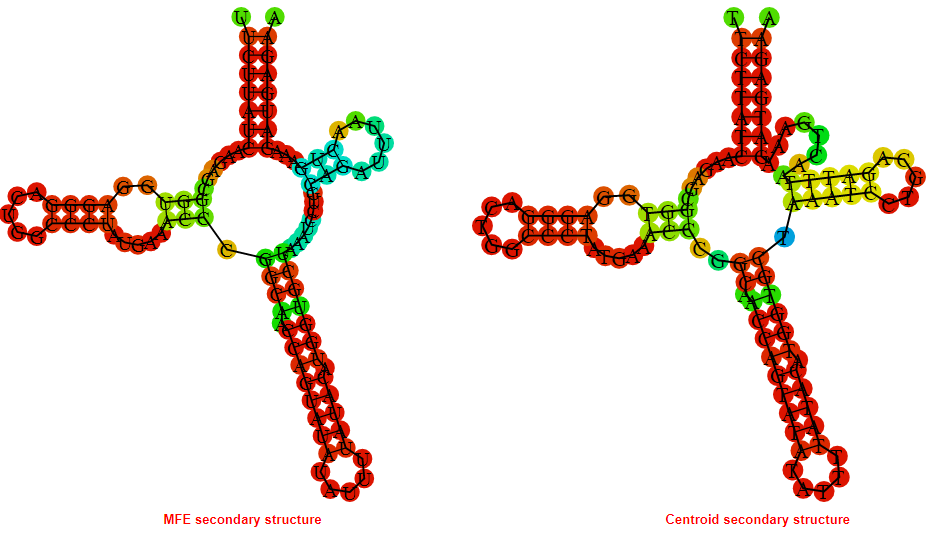
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
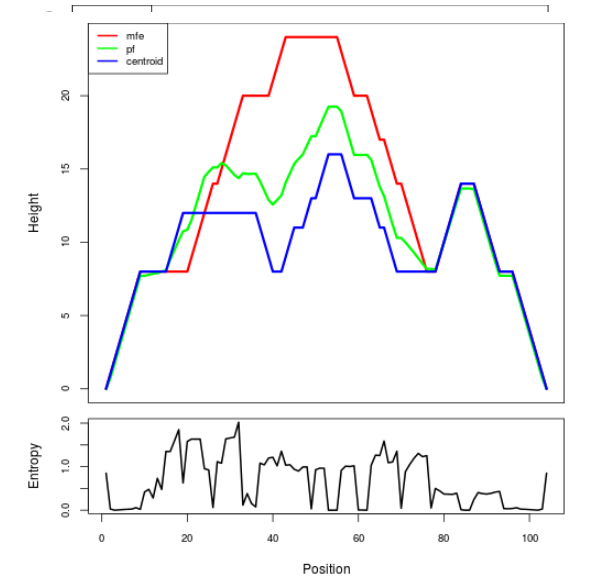
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
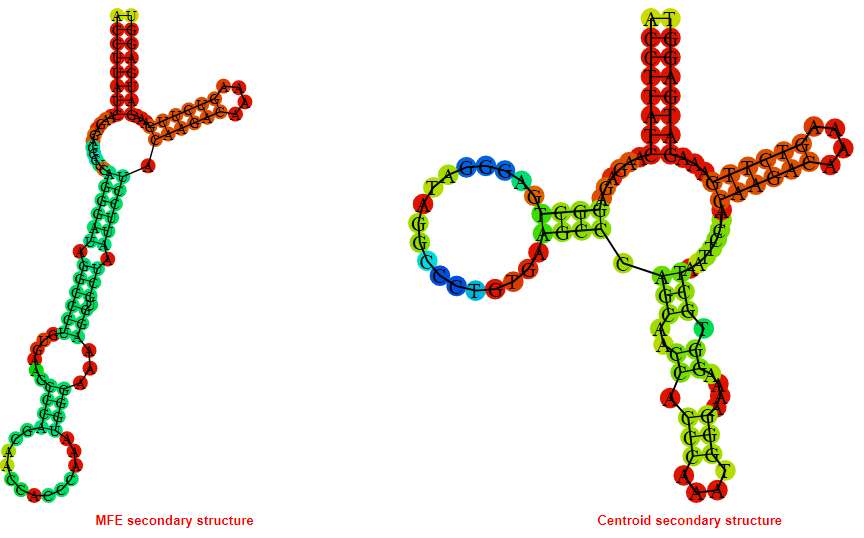
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
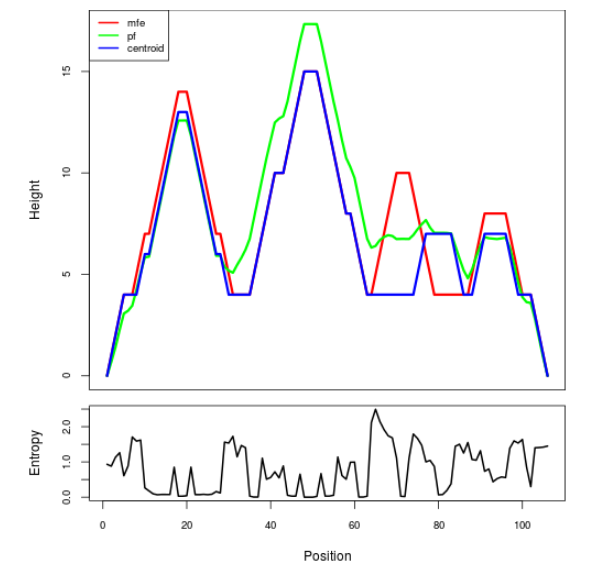
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
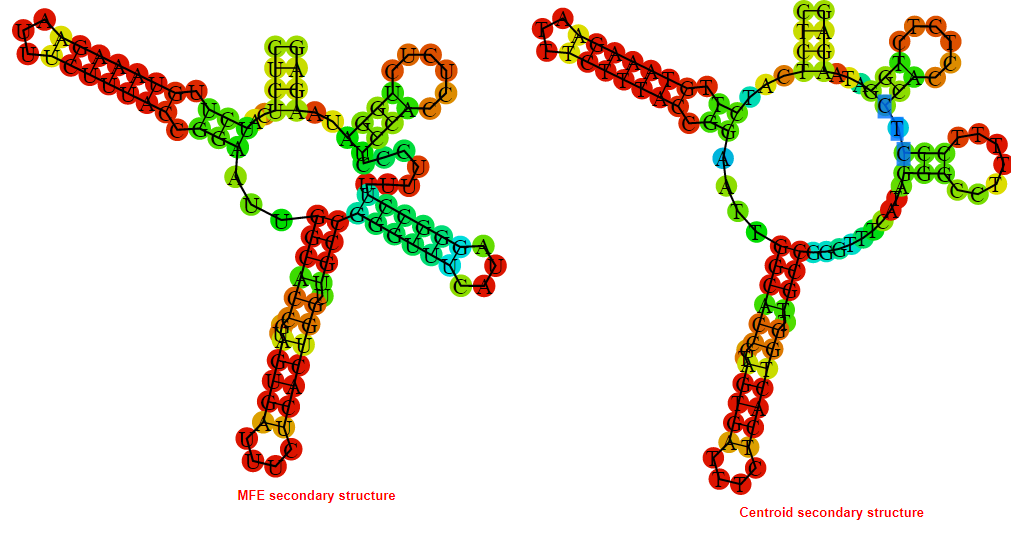
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
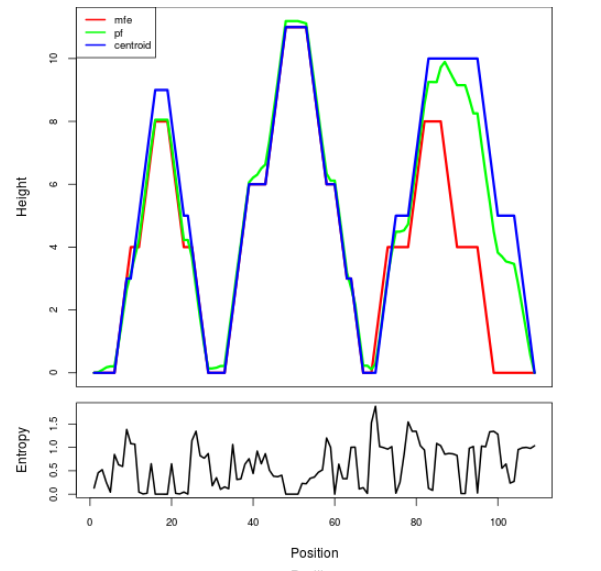
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
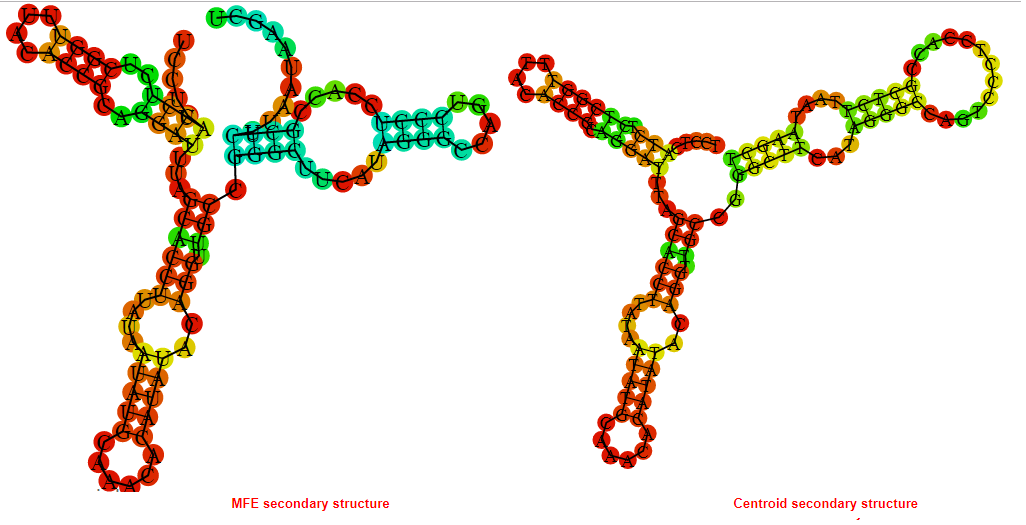
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
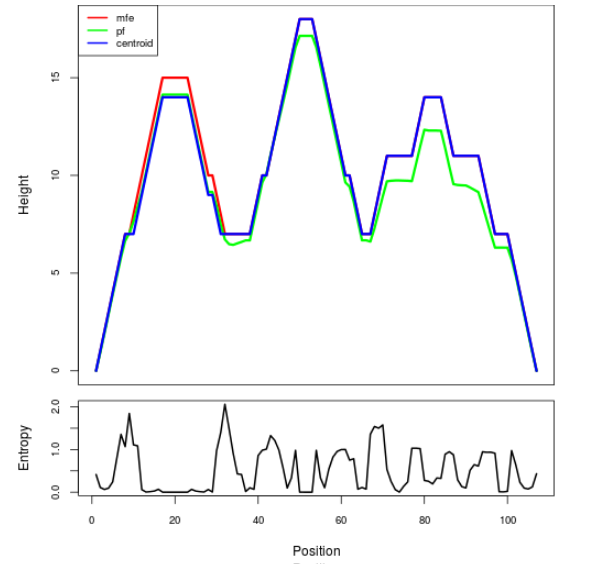
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
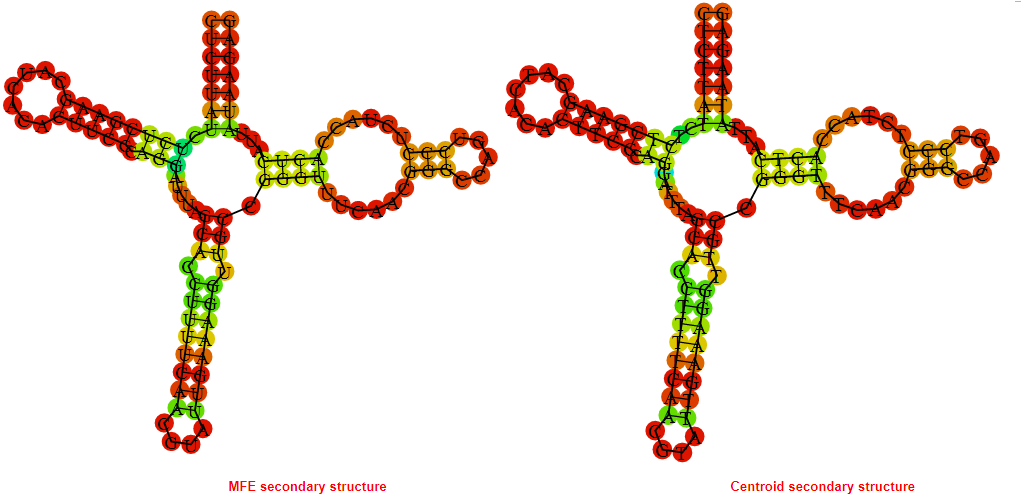
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
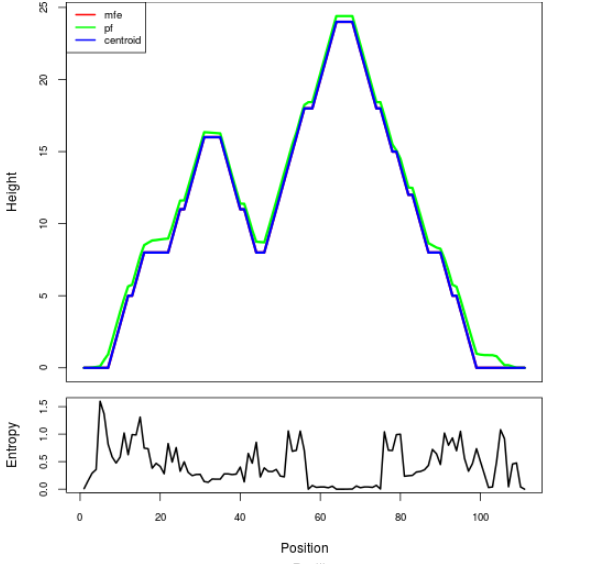
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria meningitidis, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
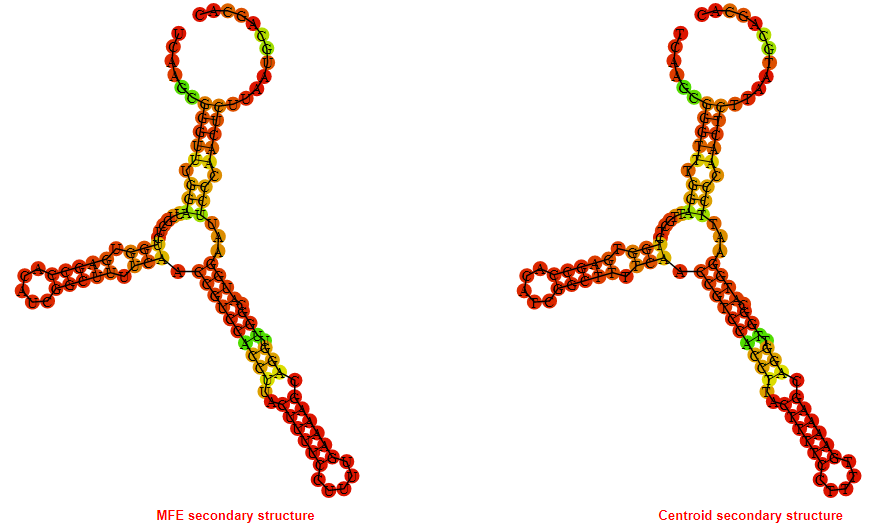
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria meningitidis, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
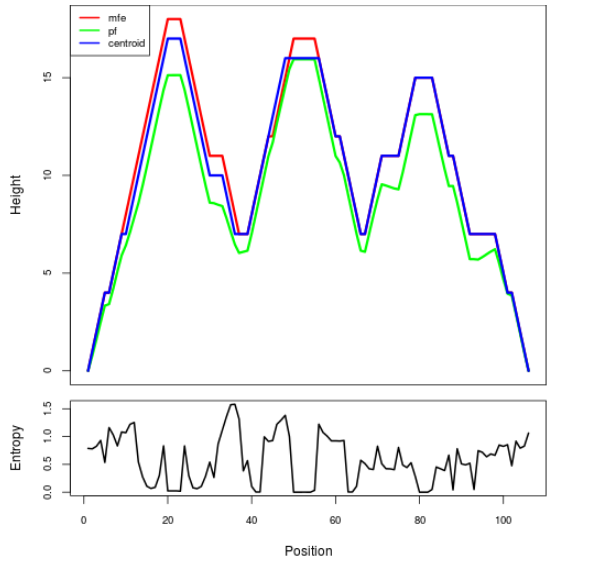
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
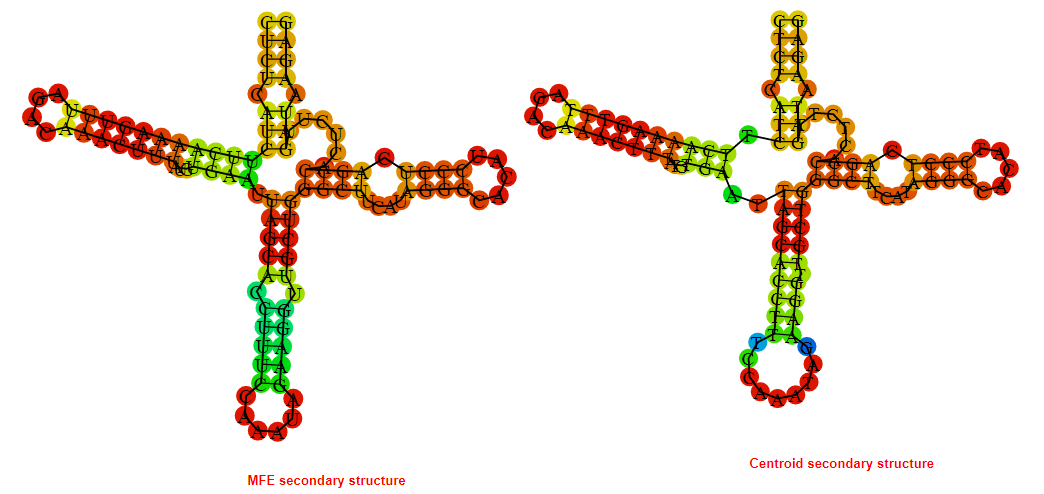
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
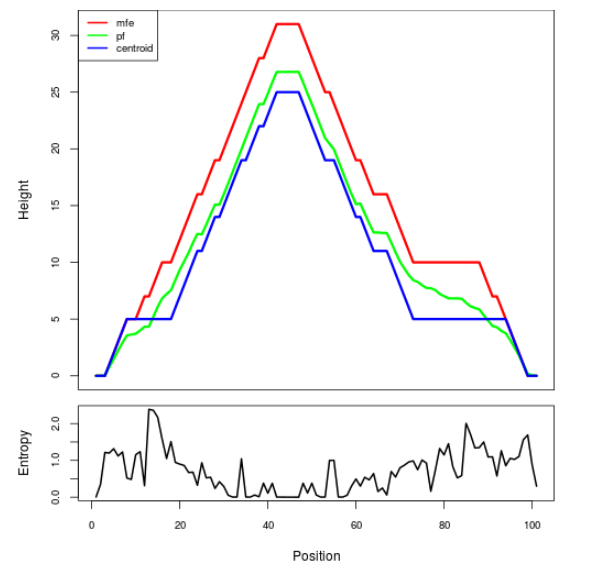
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
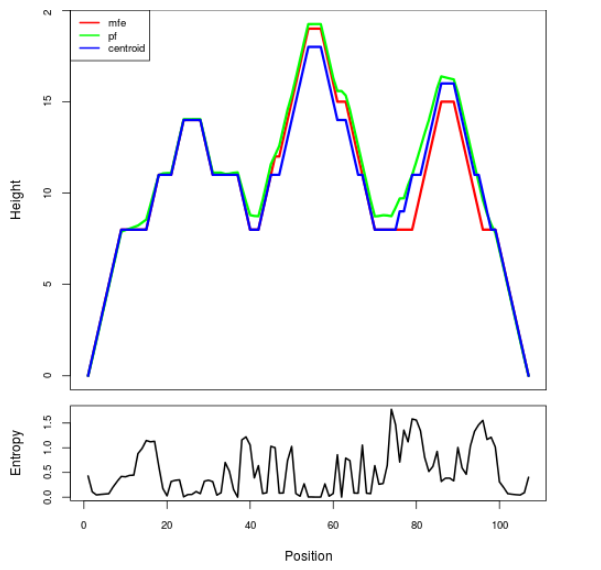
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus, SAM type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
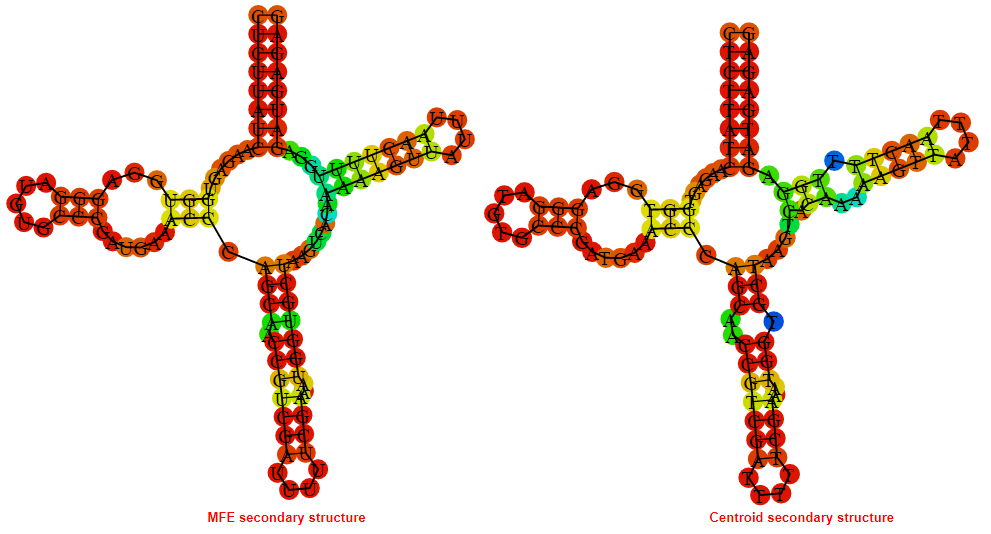
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus, SAM type switch.
Multiple alignment of the SAM type switches
Below is presented the sequences alignment of all the SAM type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the SAM type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the SAM type switches involved in.

Multi_resistance strains for the SAM type switches
The table displays known human pathogenic bacteria bearing the SAM type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
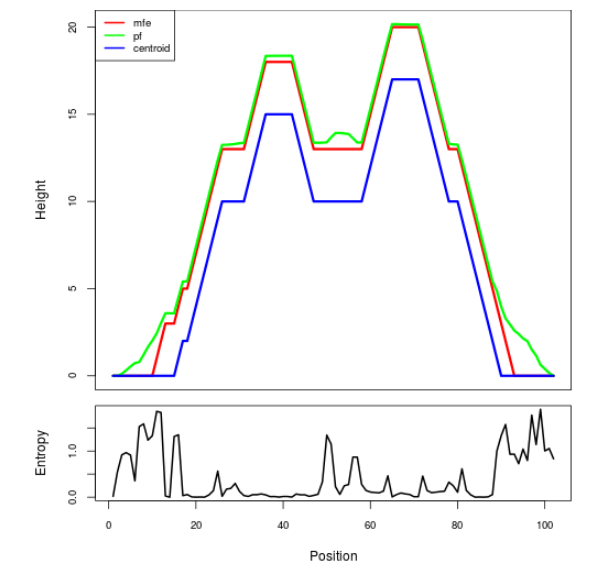
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
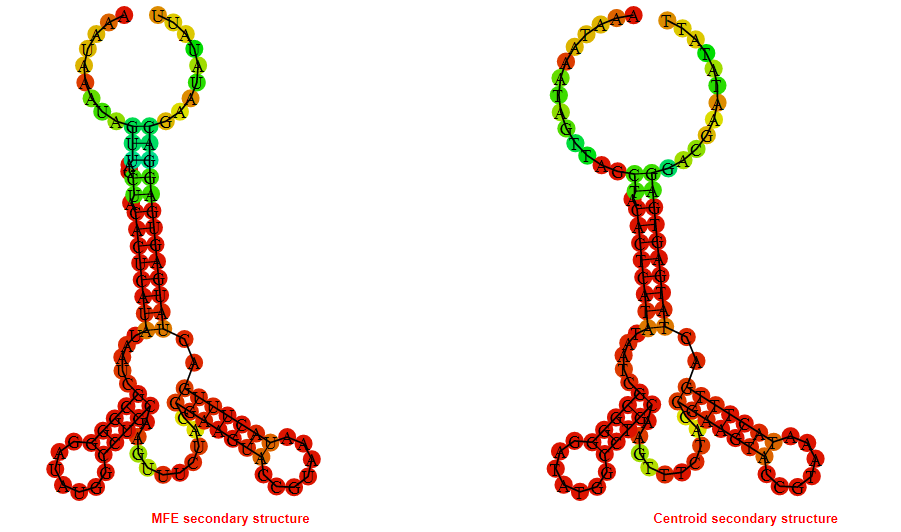
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.
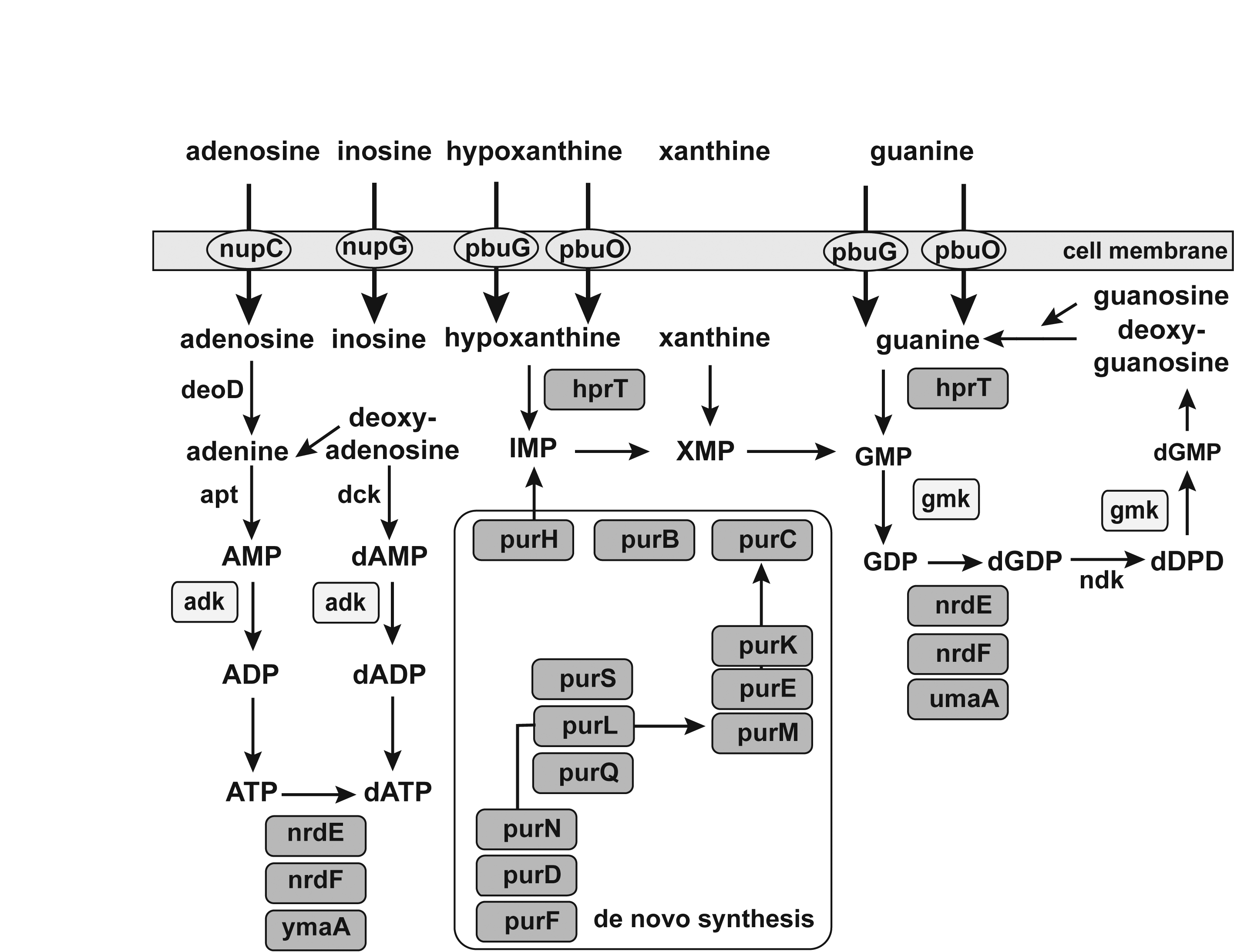
Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
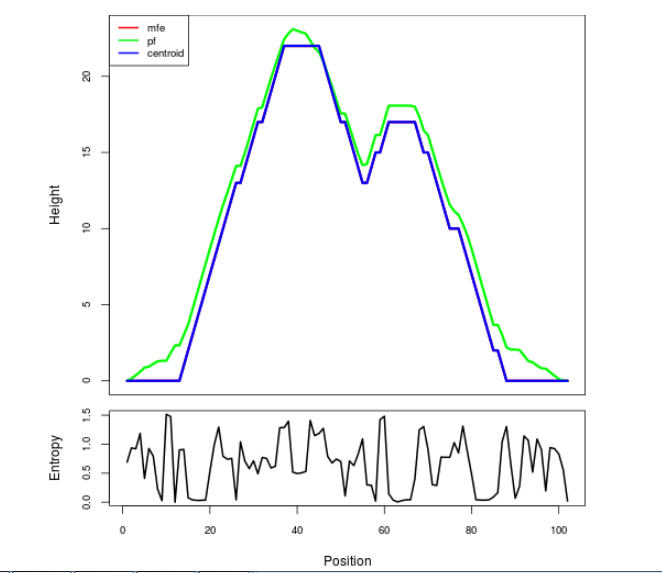
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus cereus , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
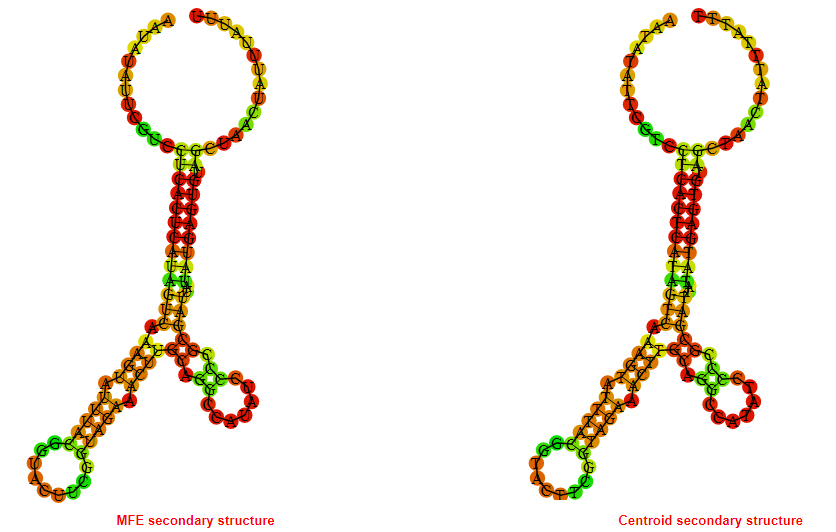
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus cereus , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
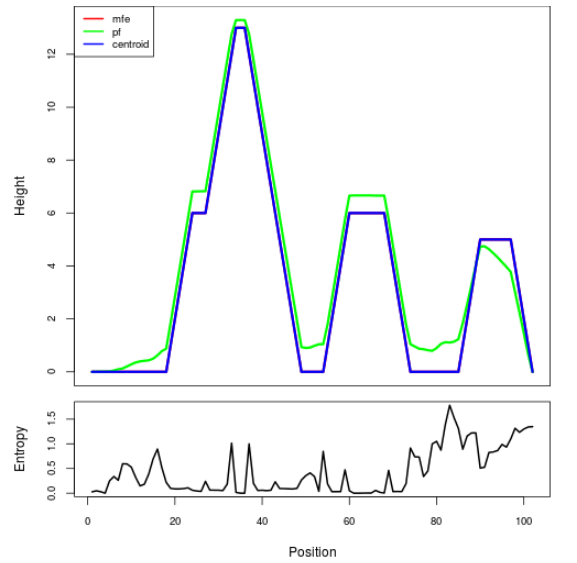
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
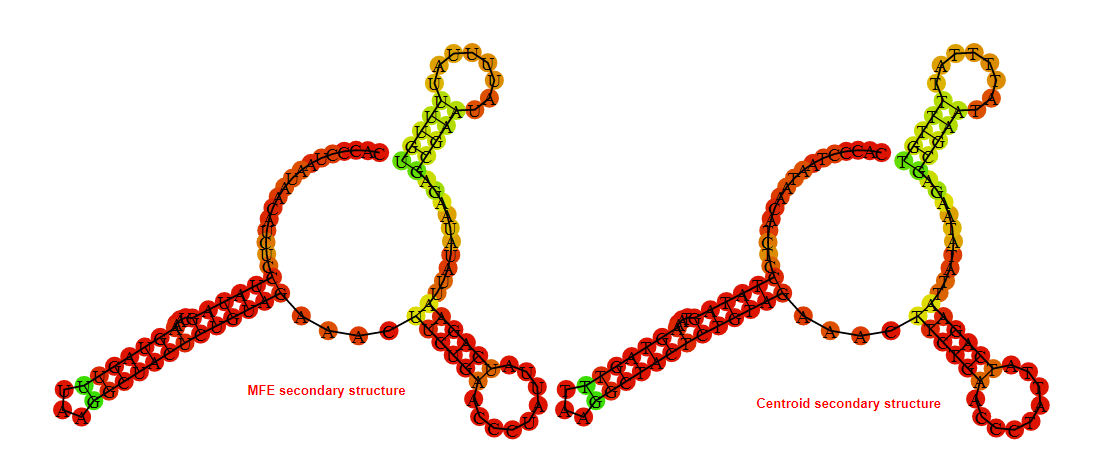
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
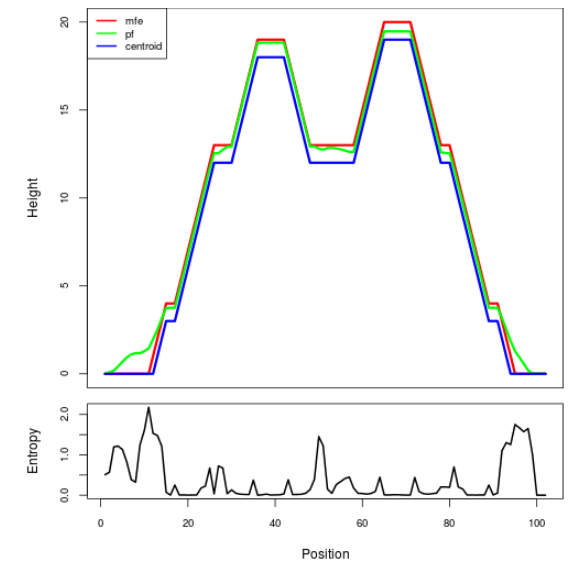
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
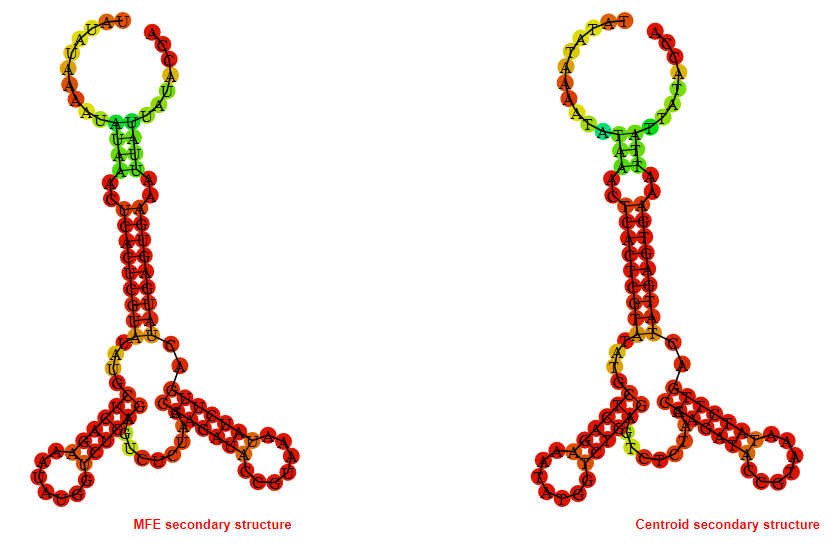
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
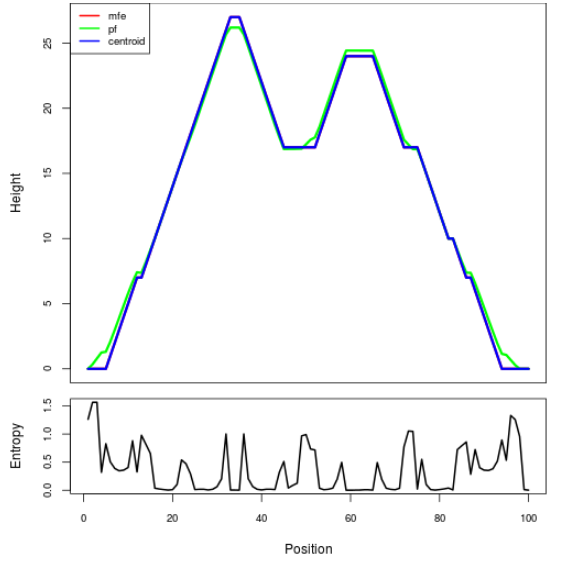
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
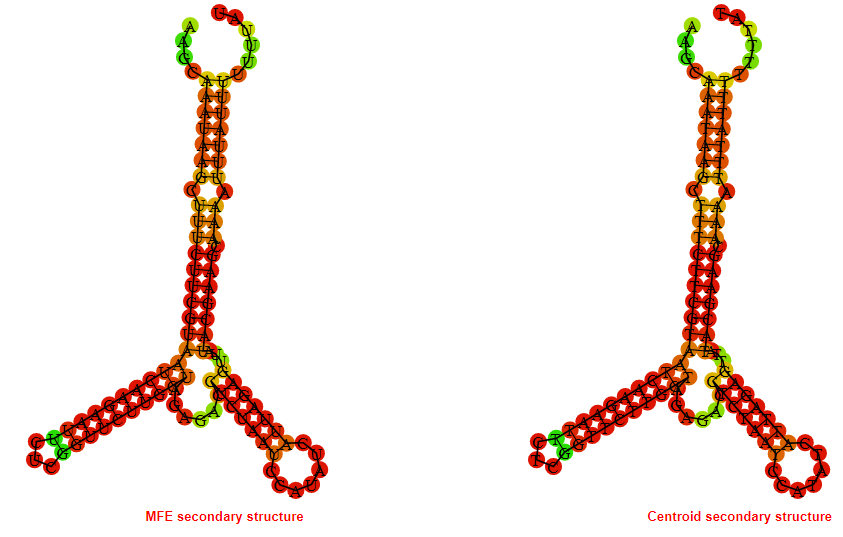
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
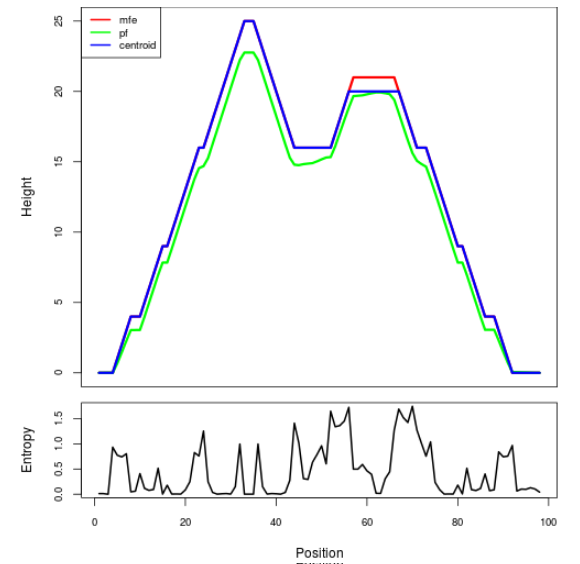
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
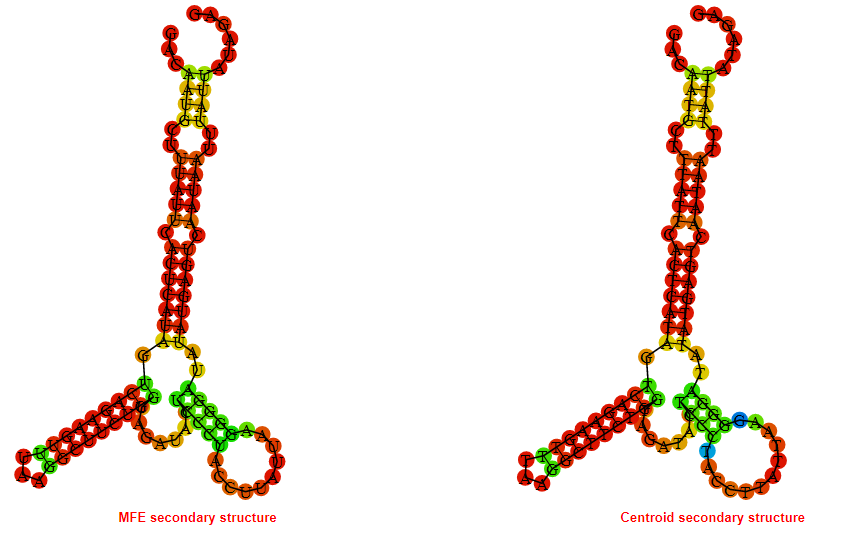
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
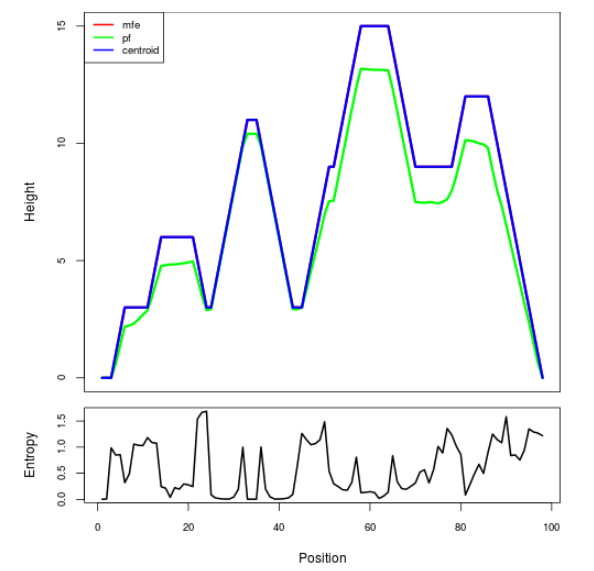
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
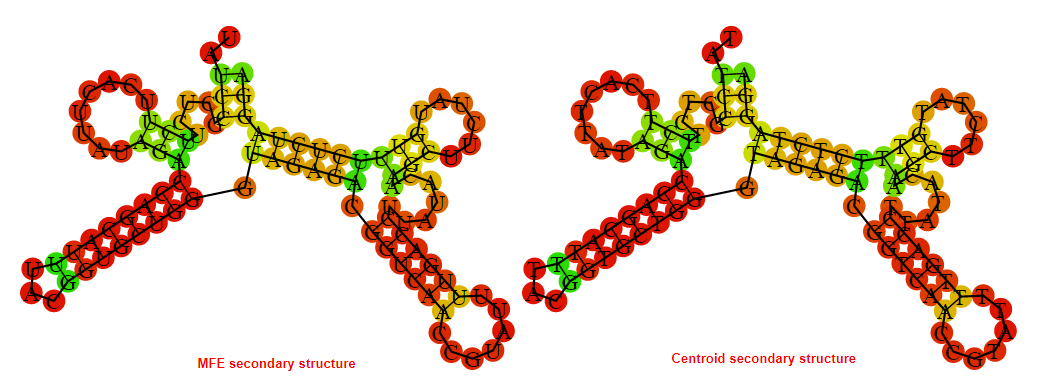
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
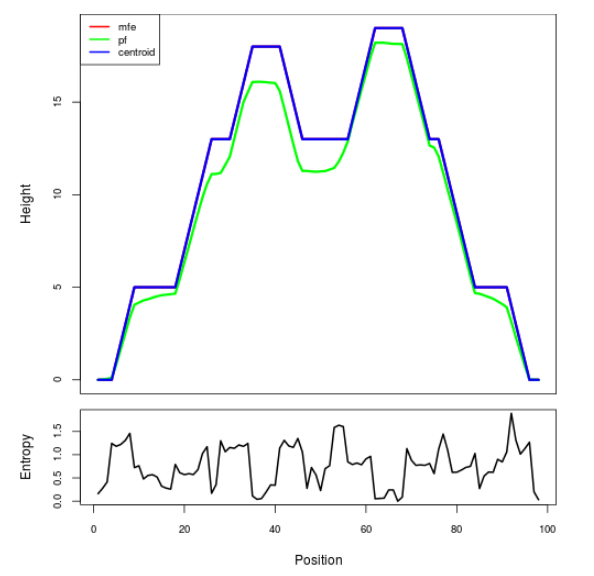
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
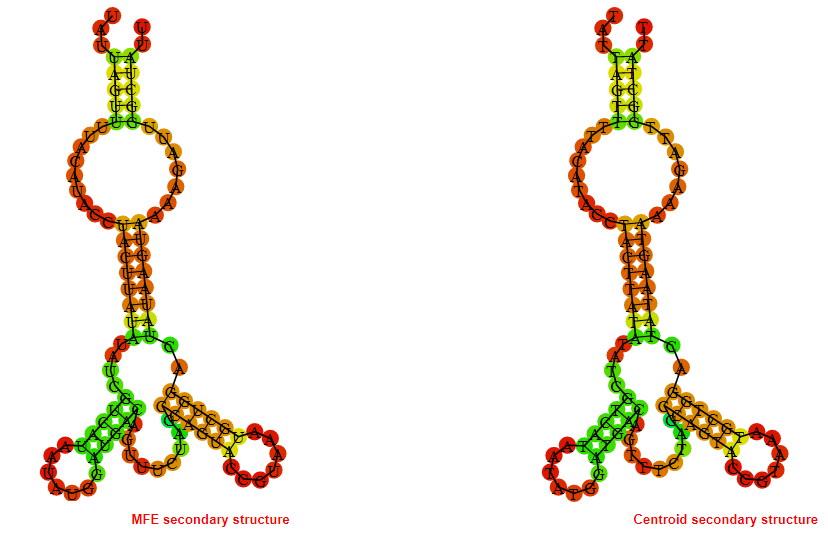
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
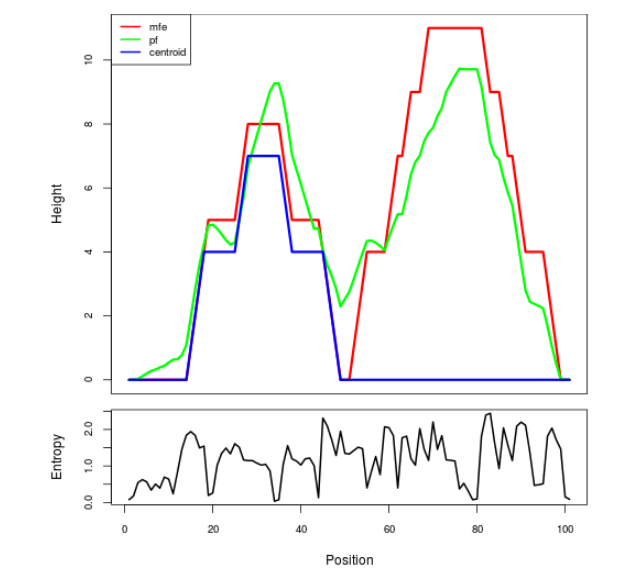
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
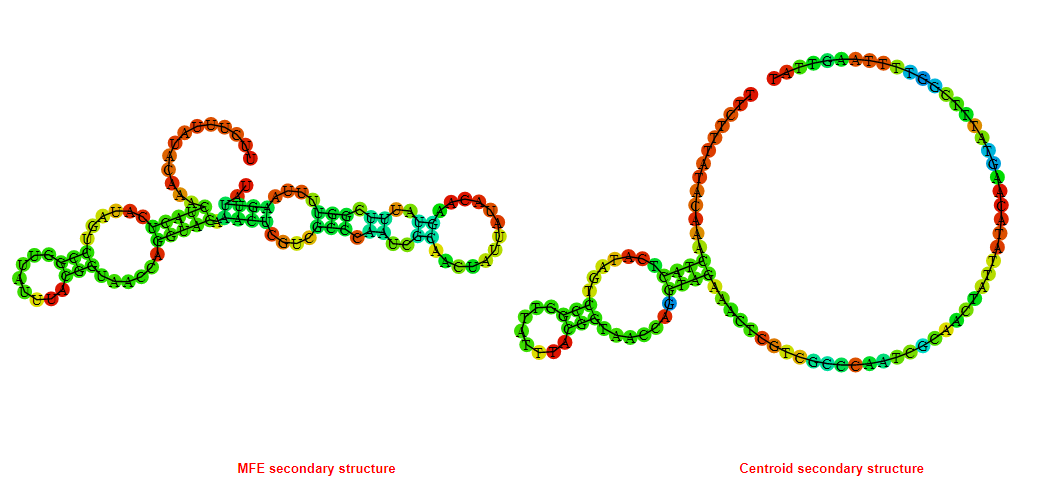
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
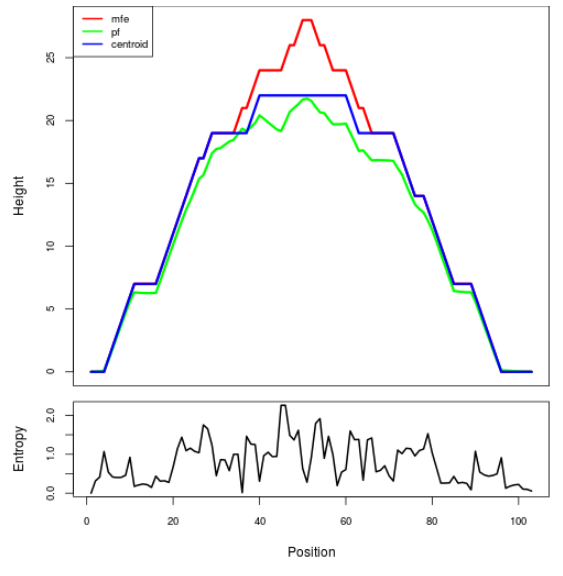
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
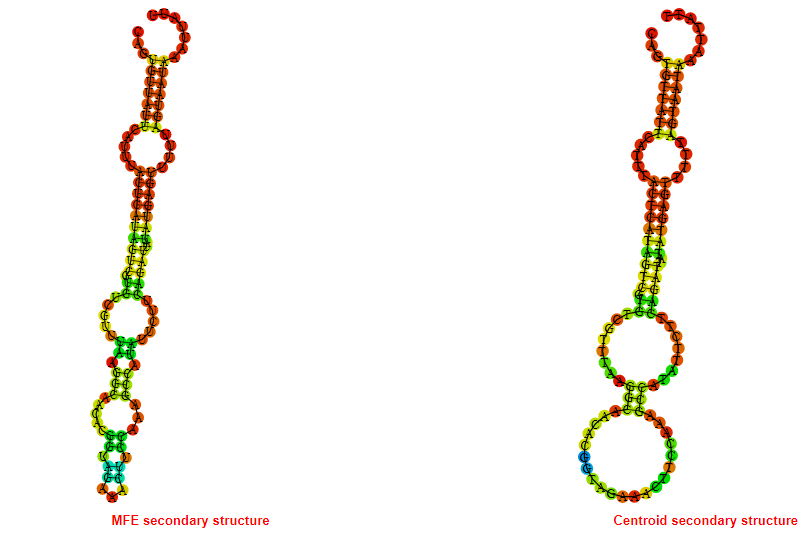
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
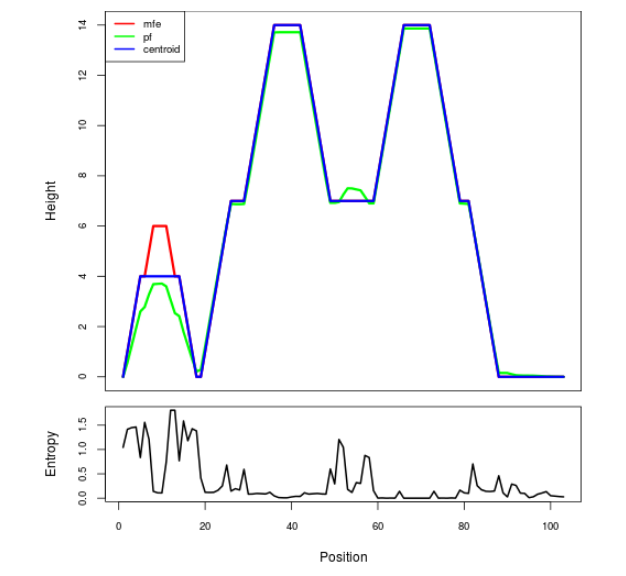
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis, Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
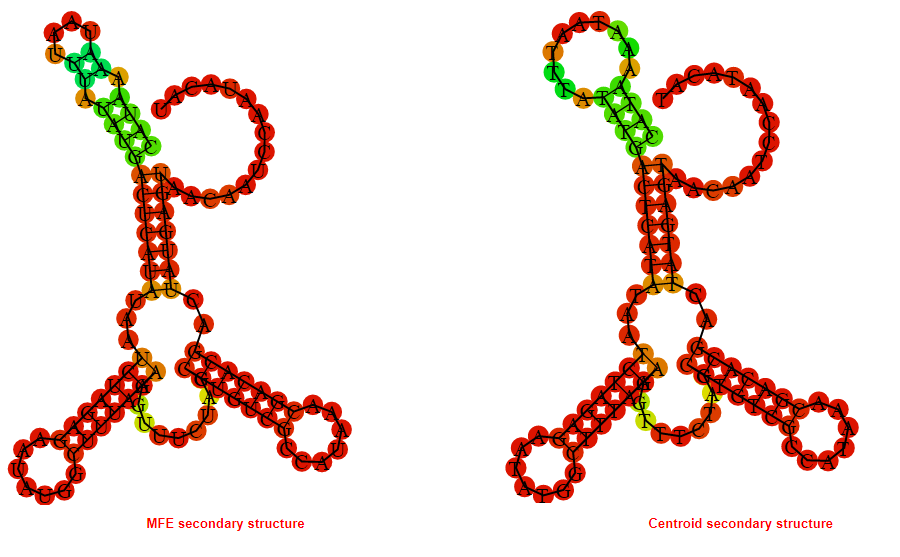
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis, Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
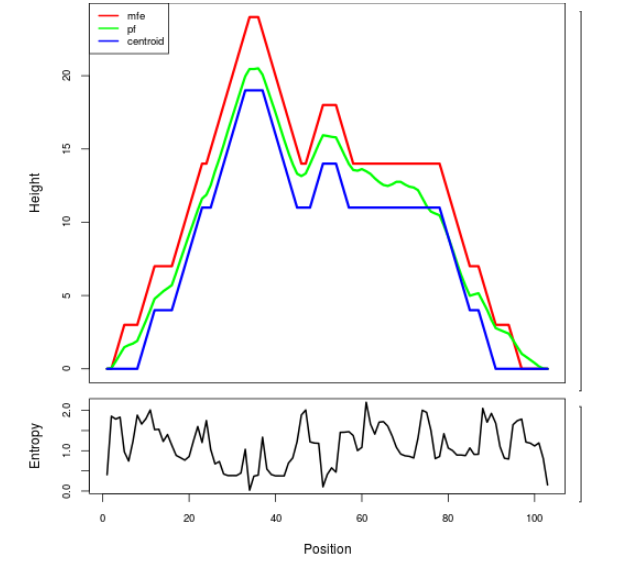
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus, Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
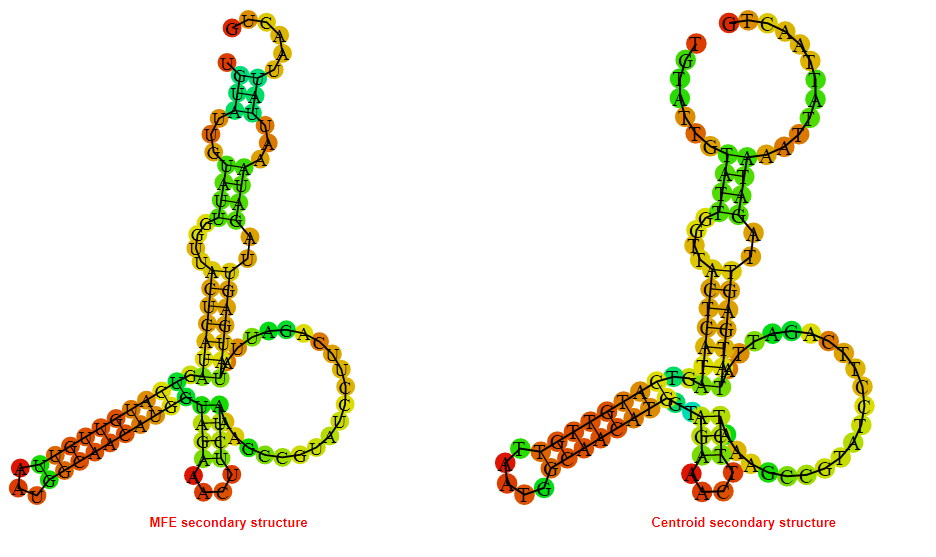
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus, Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
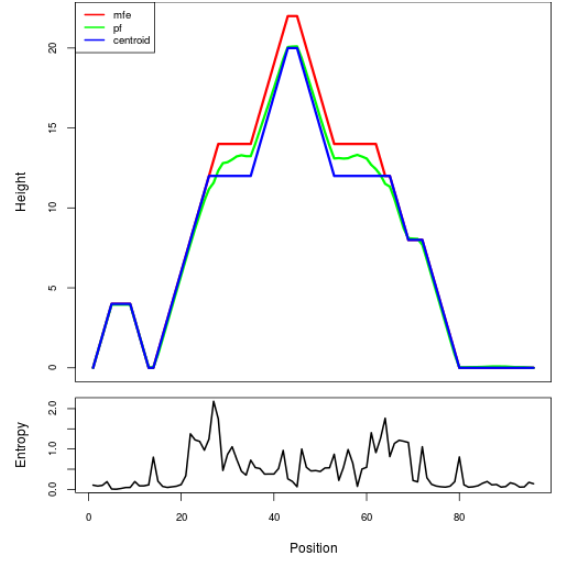
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus agalactiae, Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
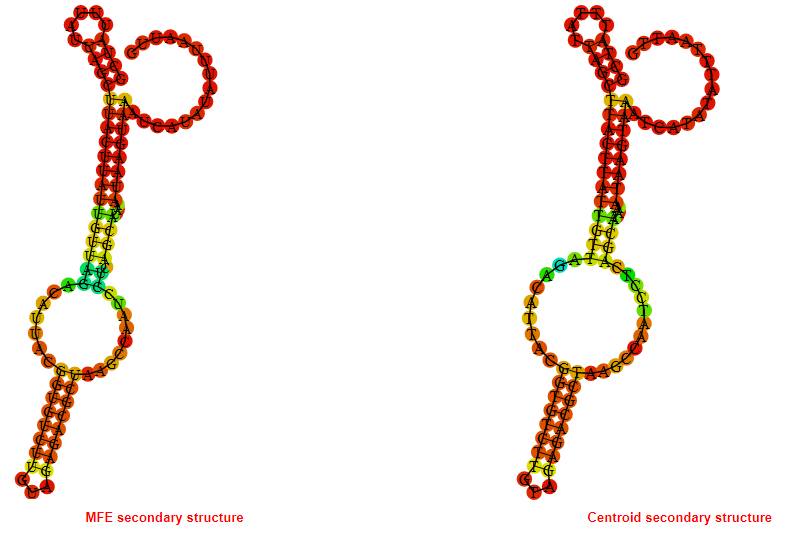
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus agalactiae, Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
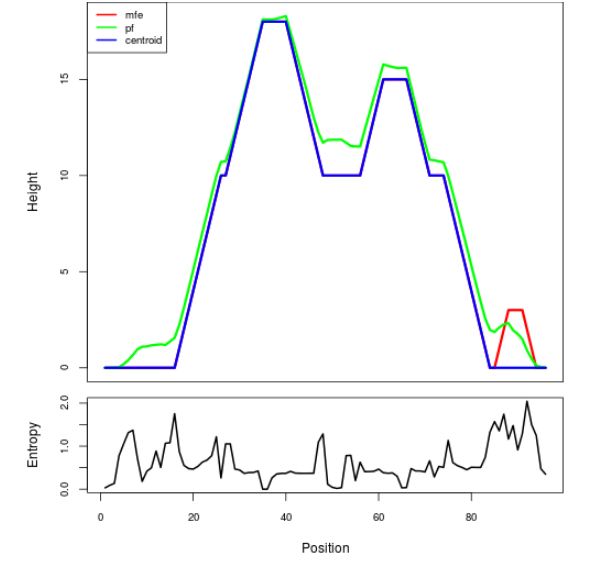
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pneumoniae , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
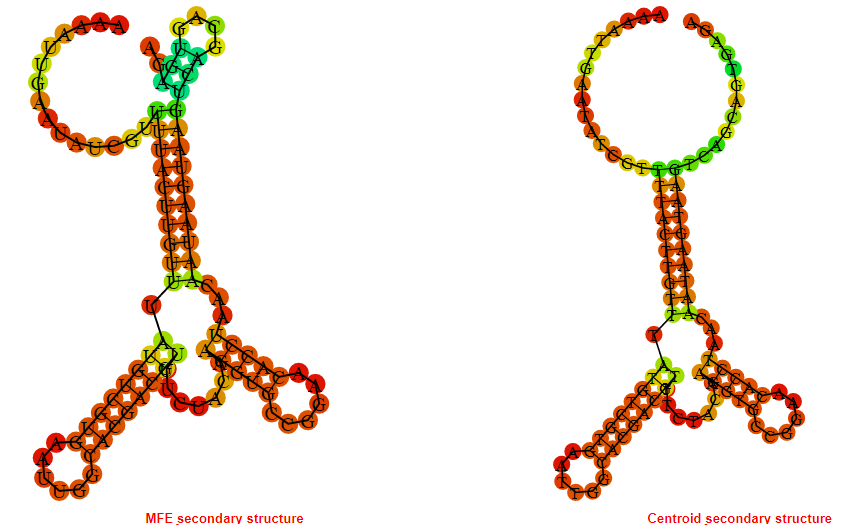
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pneumoniae , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
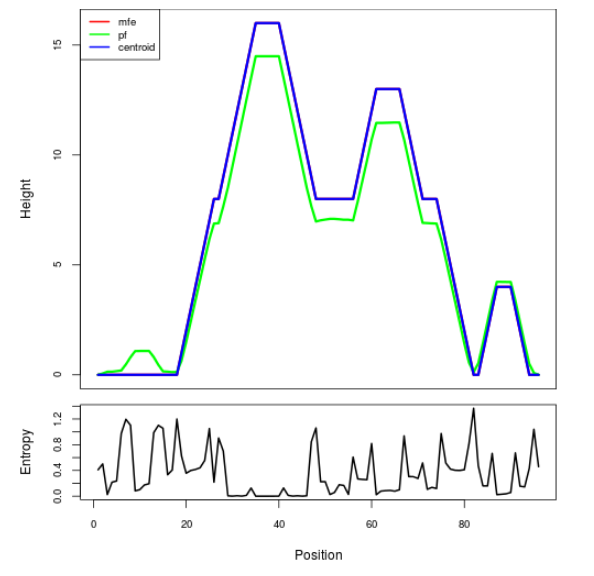
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pyogenes , Purine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
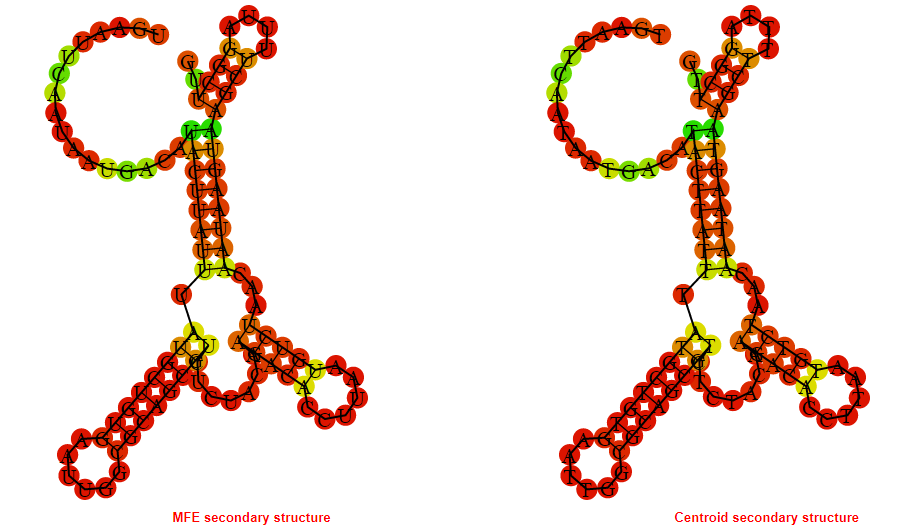
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pyogenes , Purine type switch.
Multiple alignment of the Purine type switches
Below is presented the sequences alignment of all the Purine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Purine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Purine type switches involved in.

Multi_resistance strains for the Purine type switches
The table displays known human pathogenic bacteria bearing the Purine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
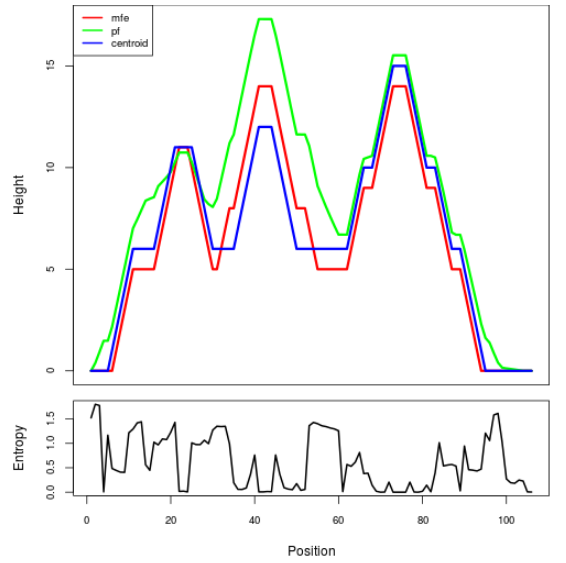
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
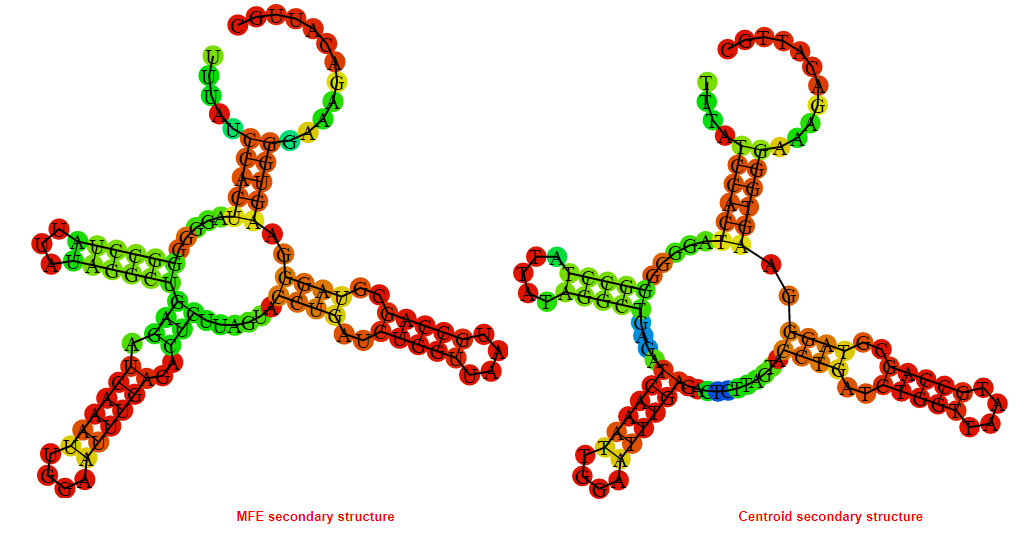
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
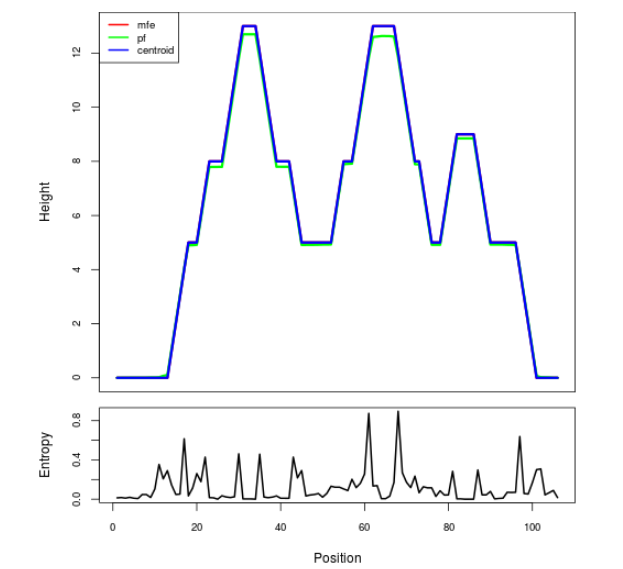
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus cereus, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
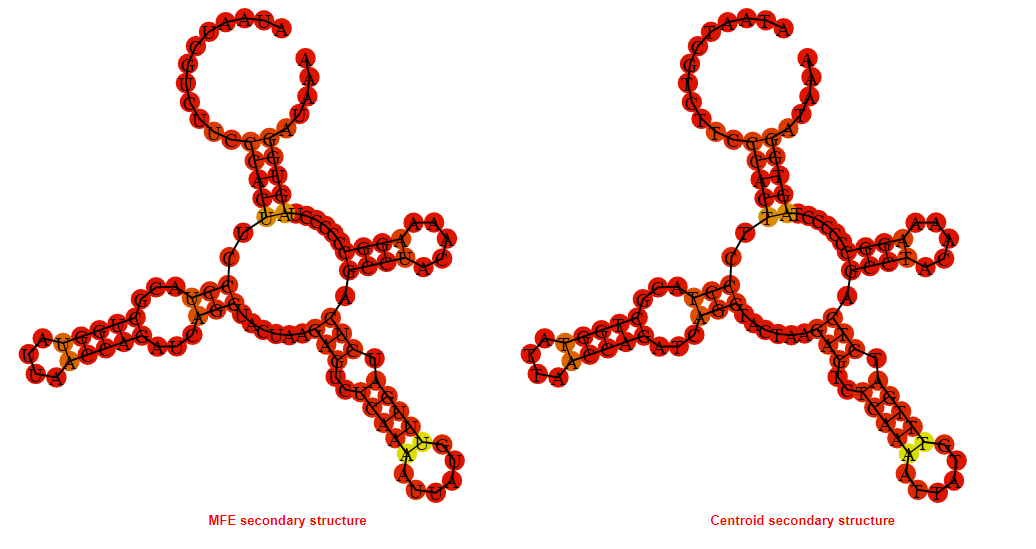
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus cereus, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
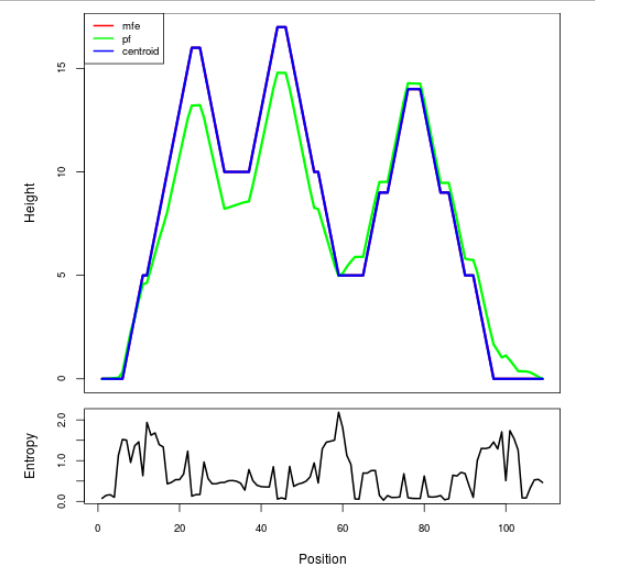
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus subtilis, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
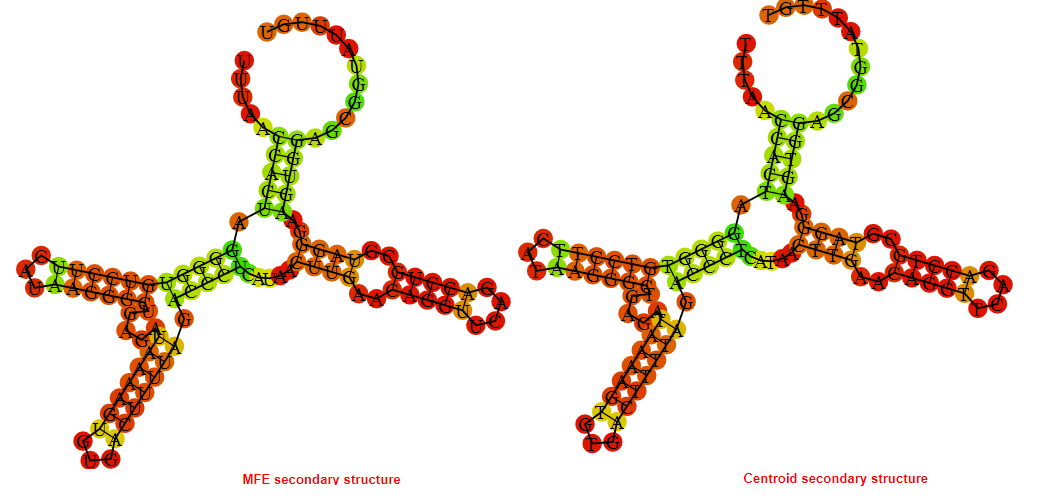
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus subtilis, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
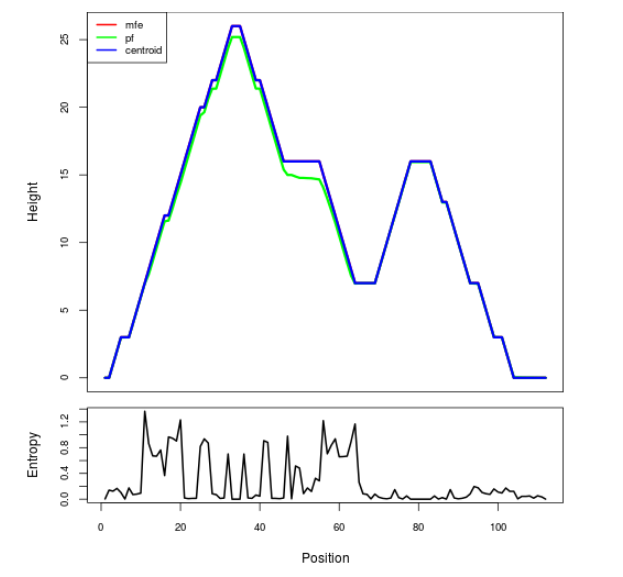
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
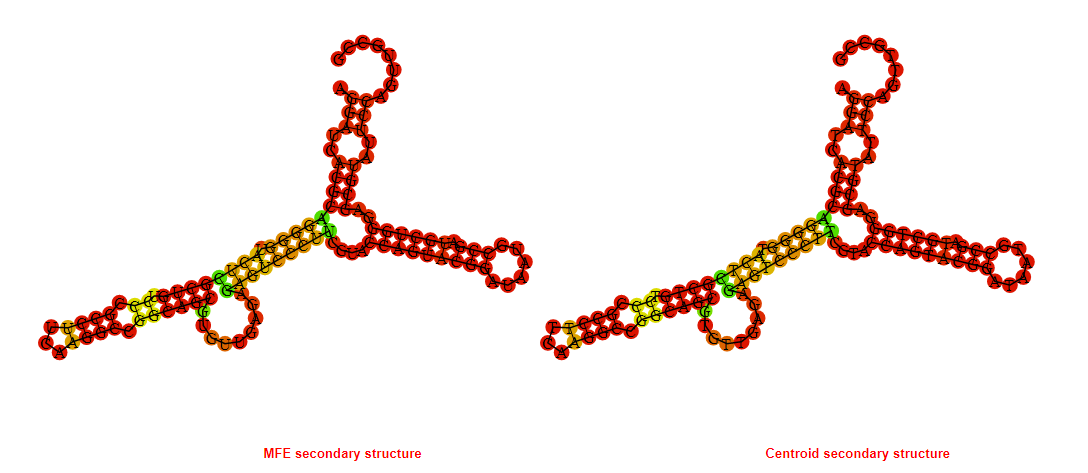
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
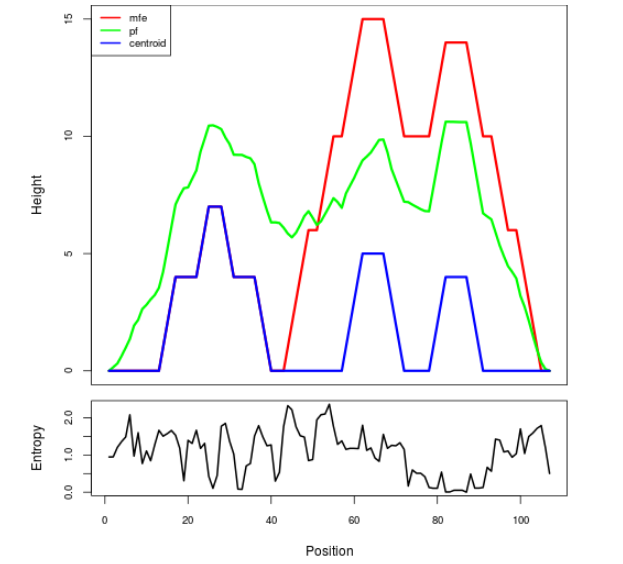
The plot represents the thermodynamic ensemble of the RNA structure of the Brucella abortus, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
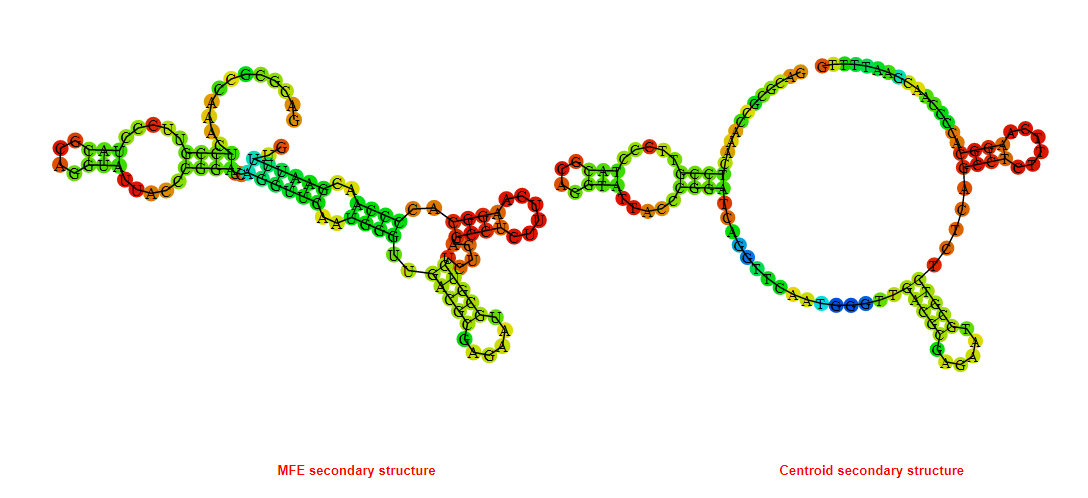
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Brucella abortus, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
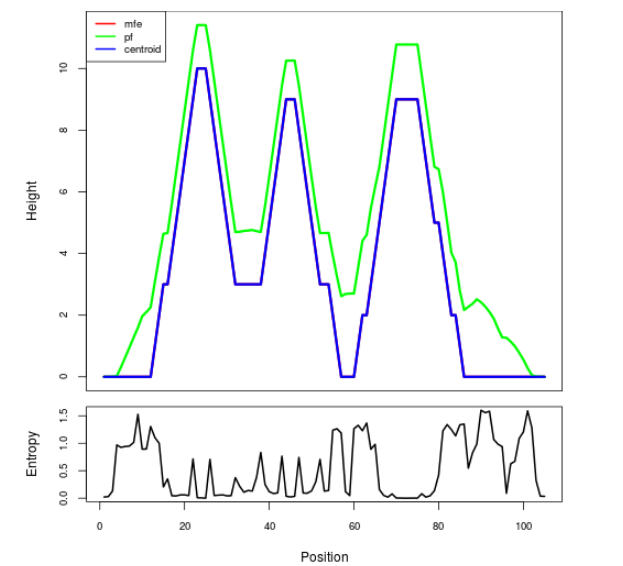
The plot represents the thermodynamic ensemble of the RNA structure of the Campylobacter jejuni, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
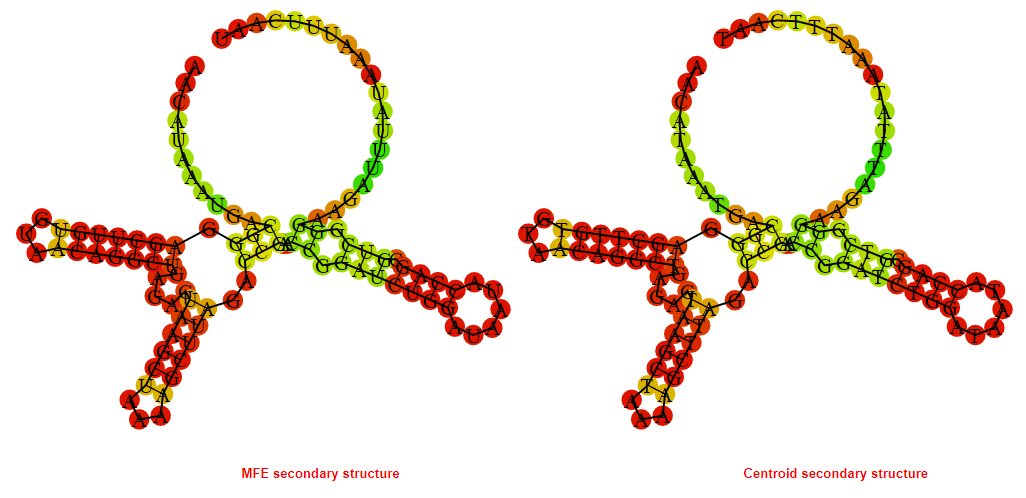
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Campylobacter jejuni, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
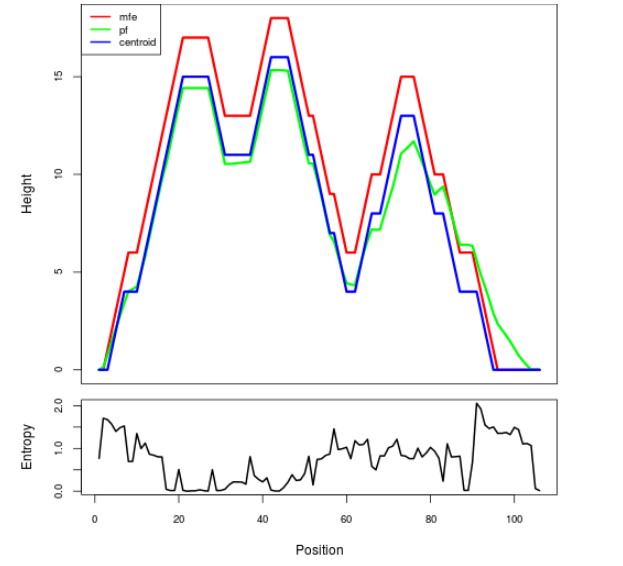
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
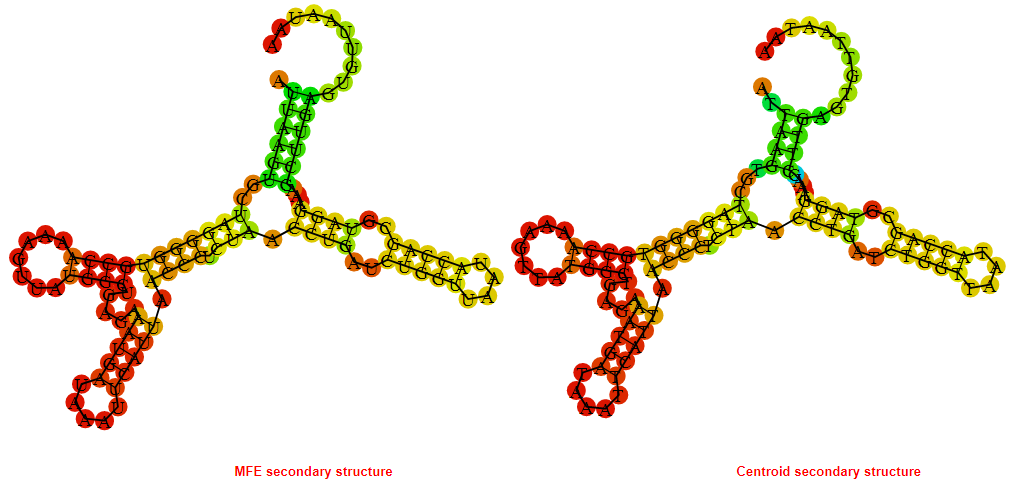
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
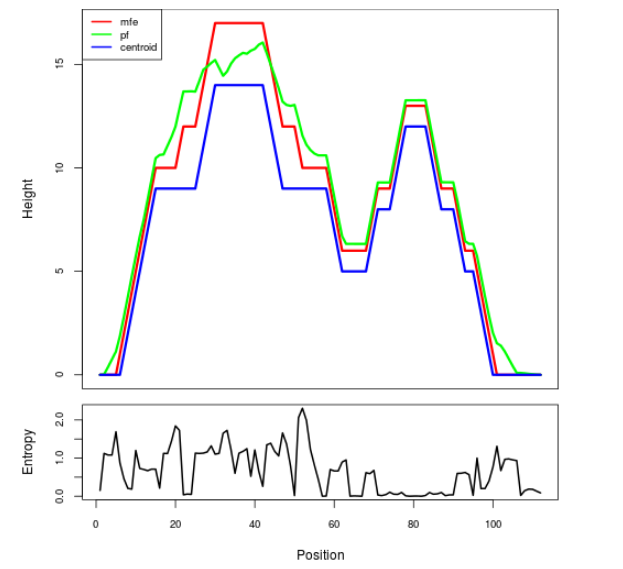
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
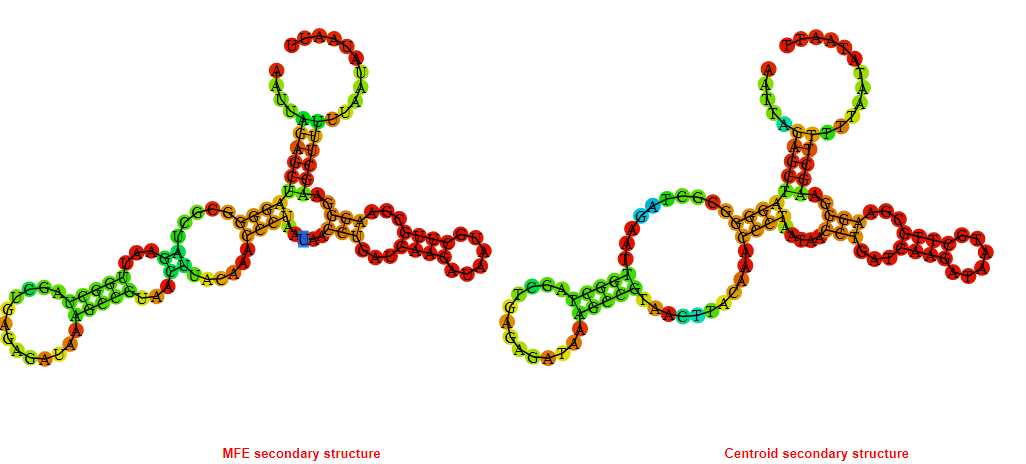
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
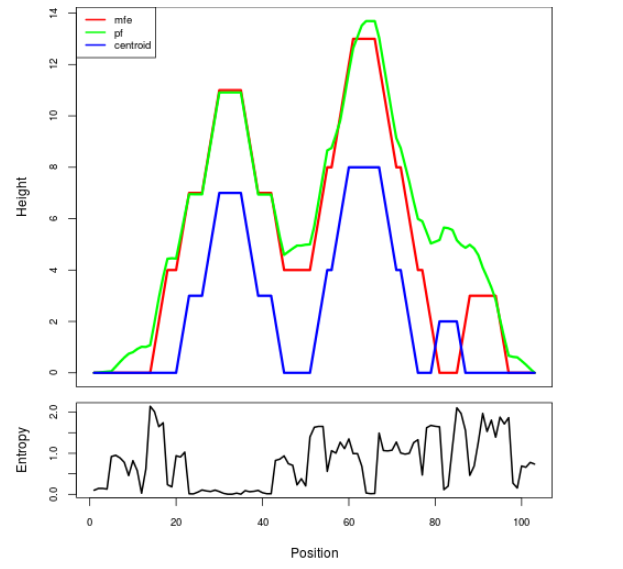
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
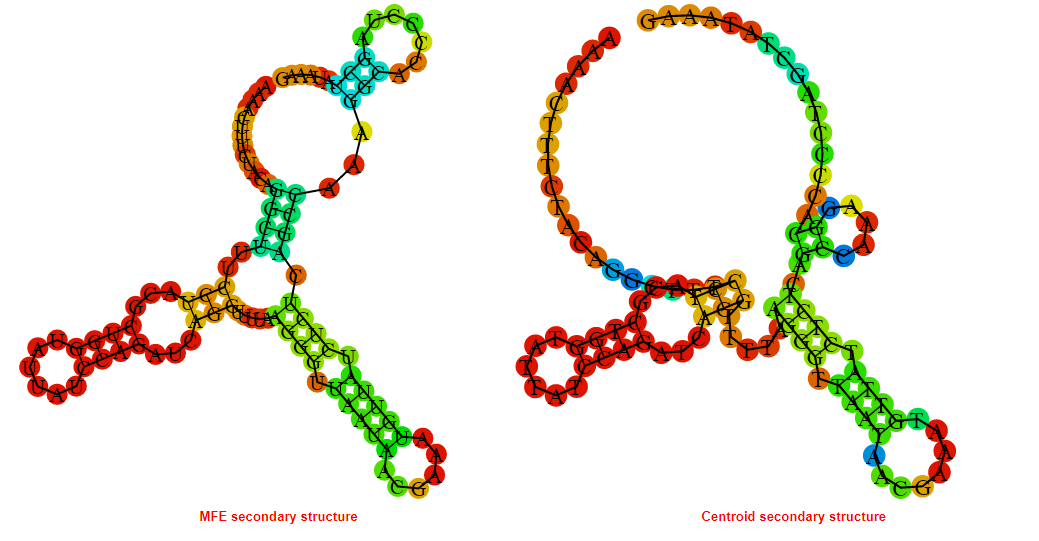
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
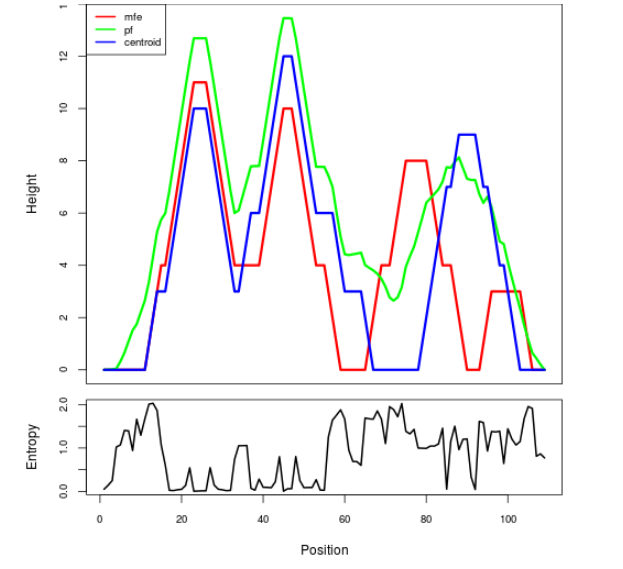
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
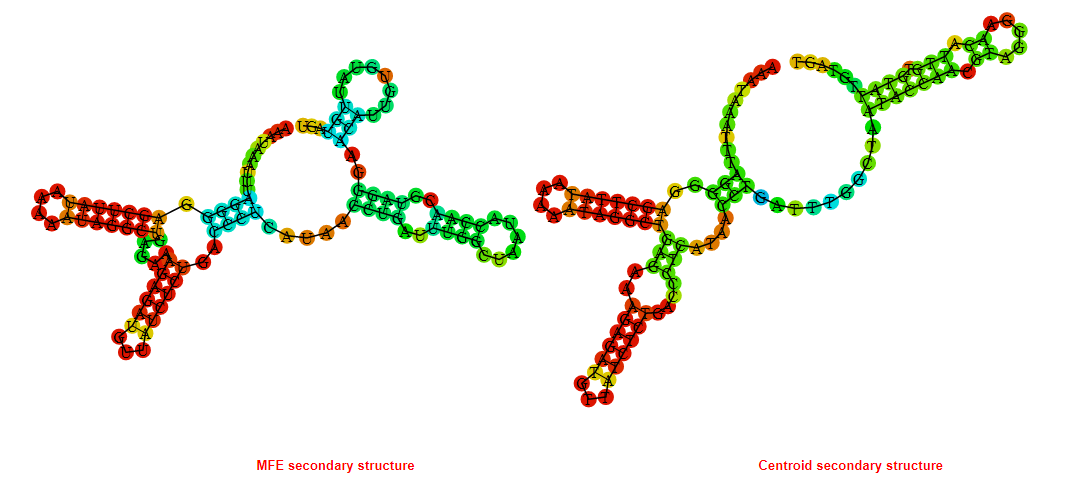
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
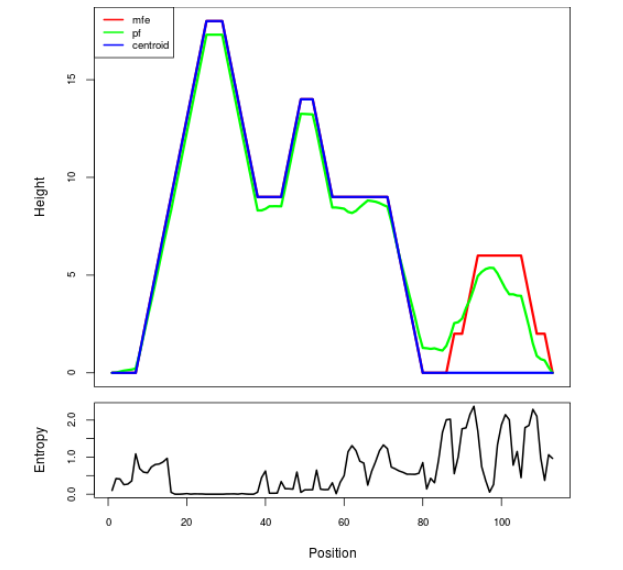
The plot represents the thermodynamic ensemble of the RNA structure of the Corynebacterium diphtheriae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
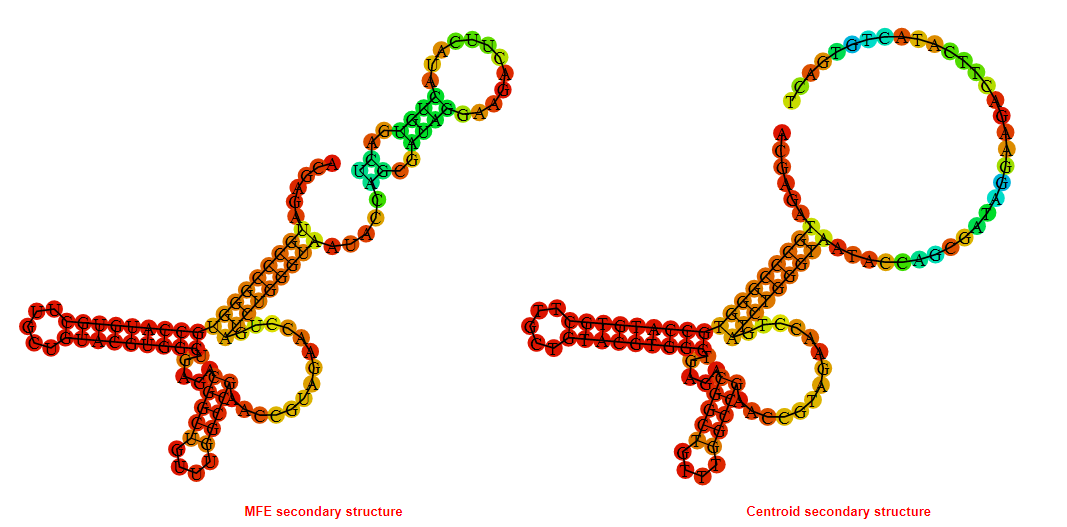
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Corynebacterium diphtheriae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
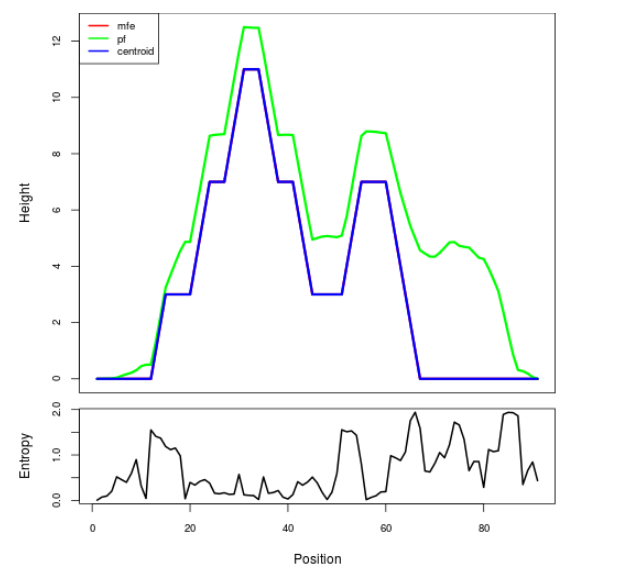
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
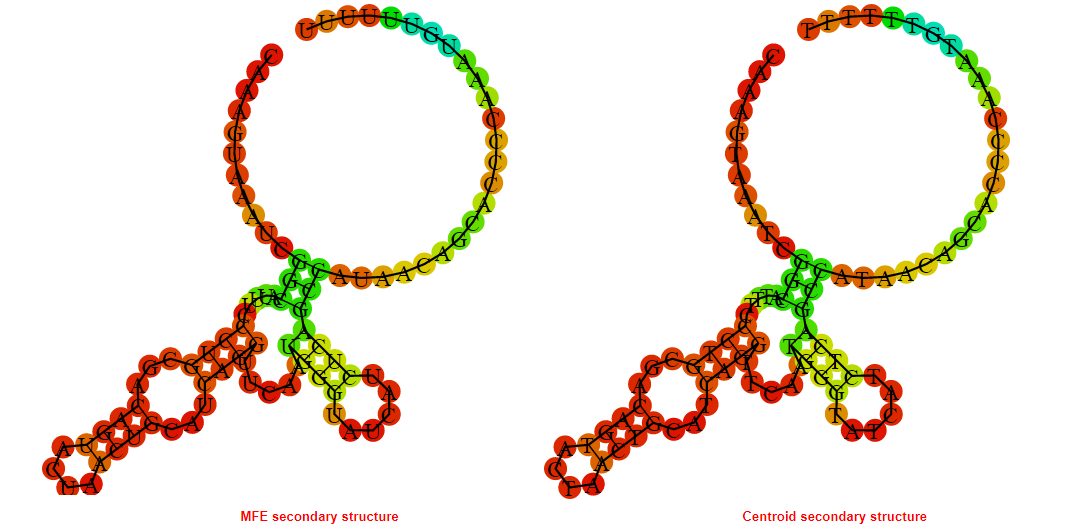
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
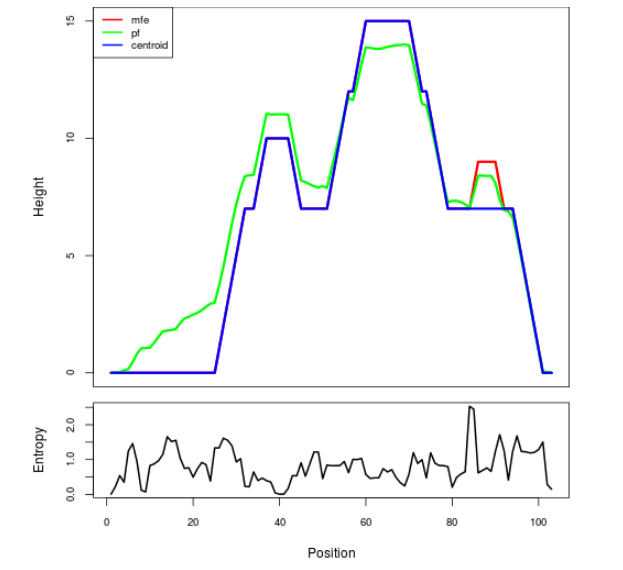
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
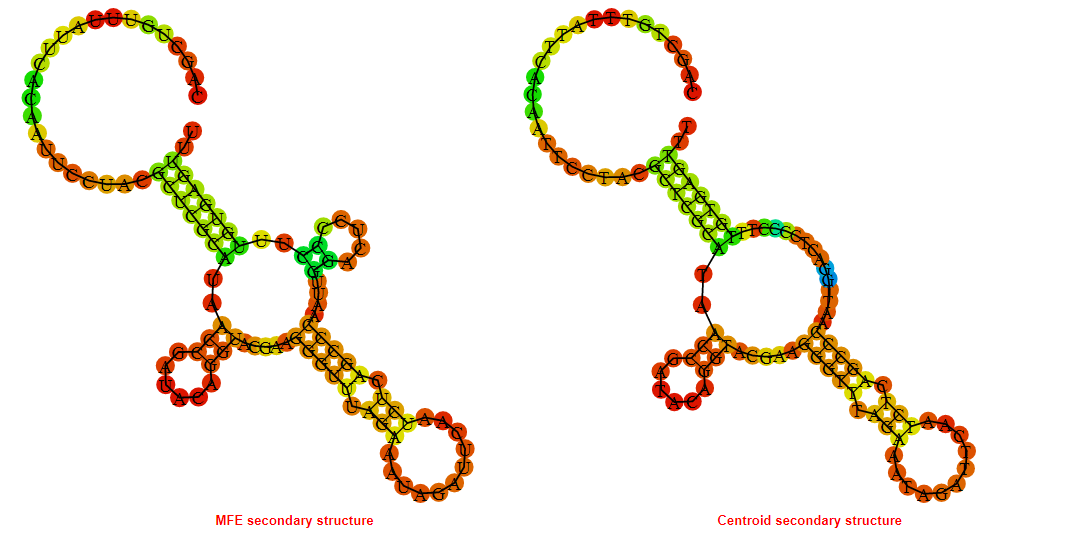
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
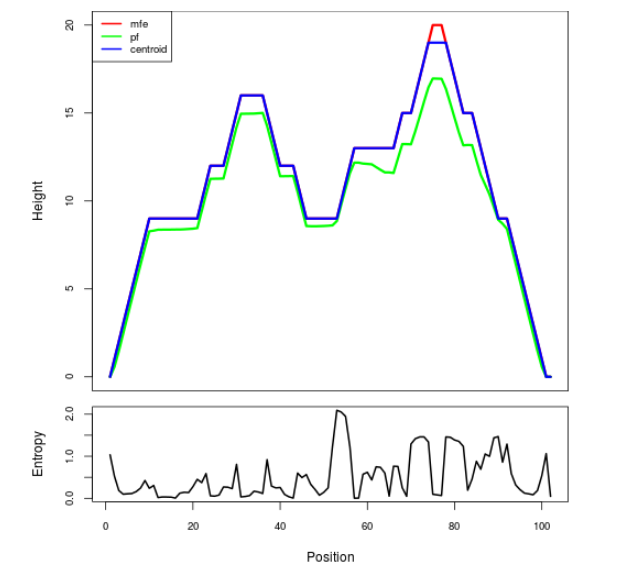
The plot represents the thermodynamic ensemble of the RNA structure of the Francisella tularensis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Francisella tularensis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
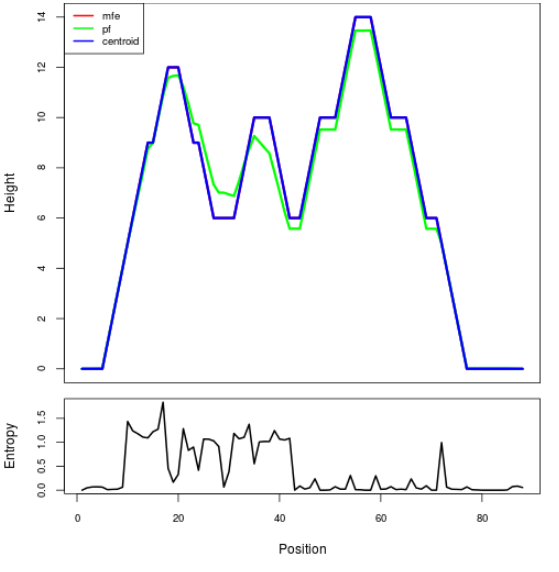
The plot represents the thermodynamic ensemble of the RNA structure of the Haemophilus influenzae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Haemophilus influenzae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
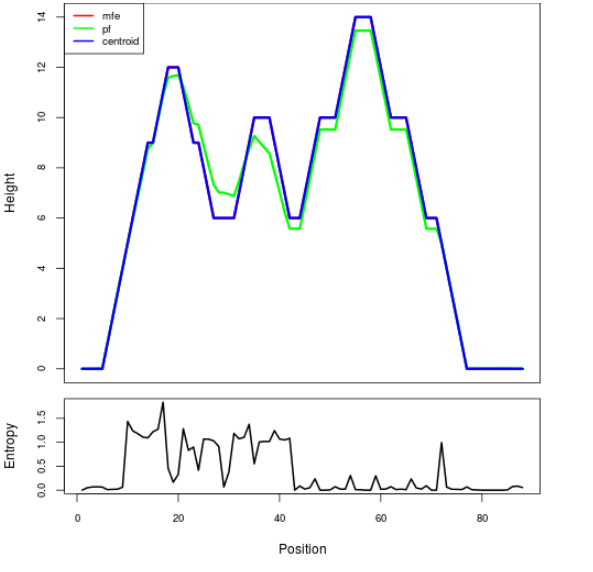
The plot represents the thermodynamic ensemble of the RNA structure of the Helicobacter pylori , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
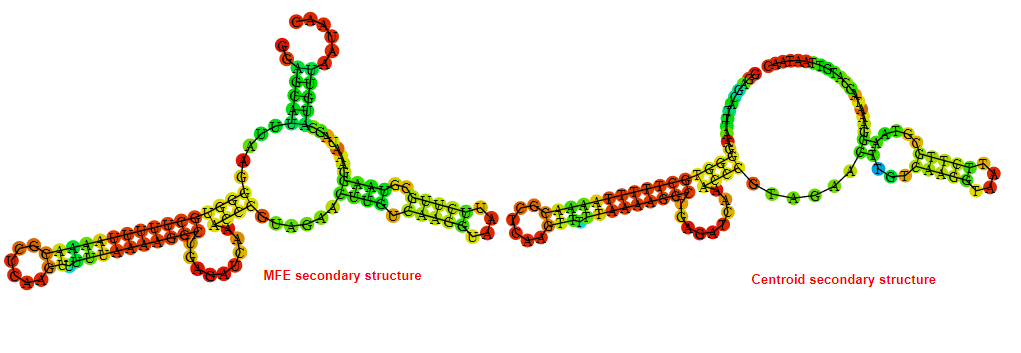
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Helicobacter pylori , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
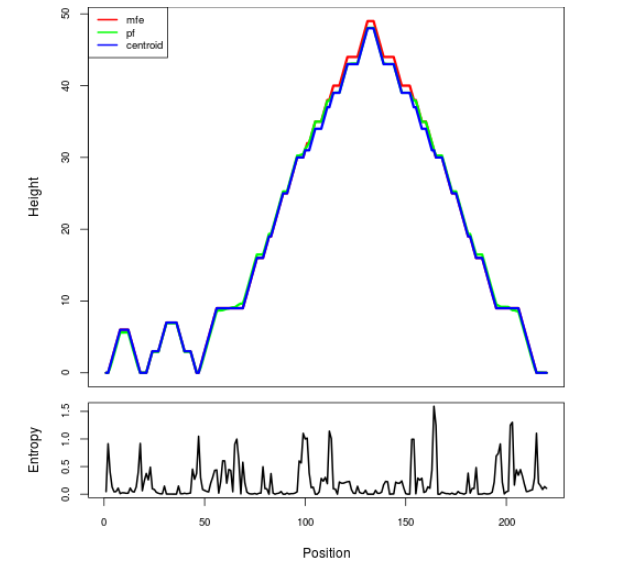
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
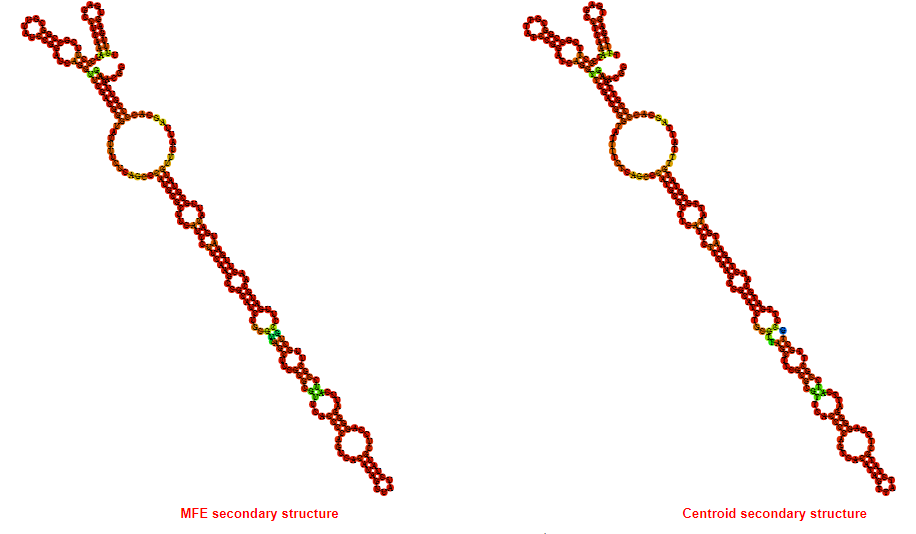
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
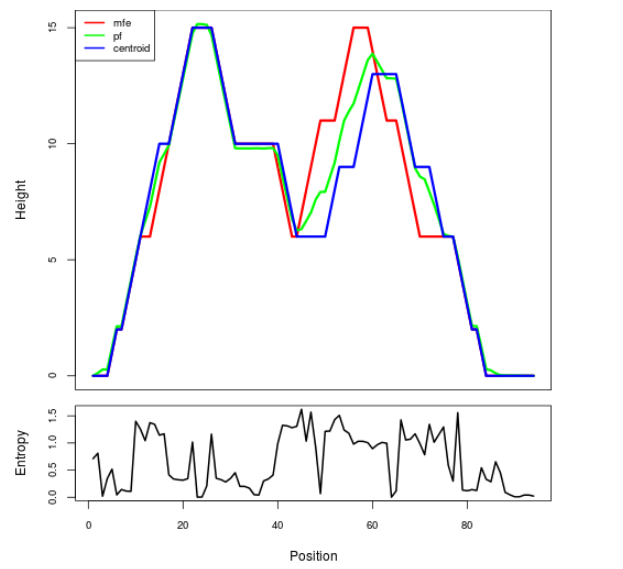
The plot represents the thermodynamic ensemble of the RNA structure of the Legionella pneumophila , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Legionella pneumophila , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
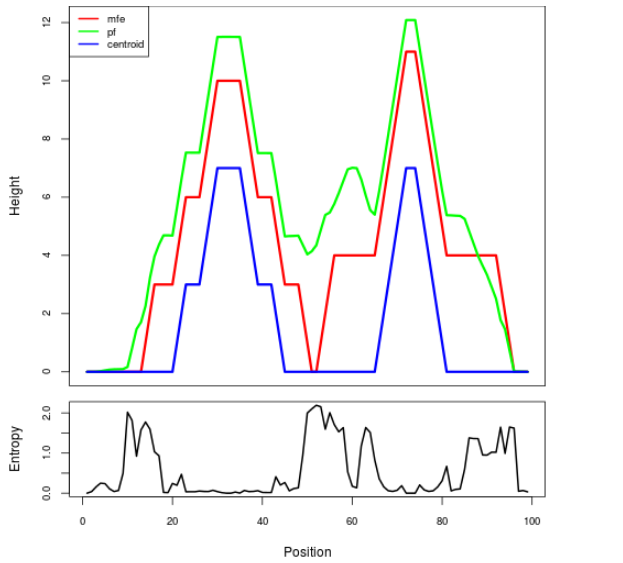
The plot represents the thermodynamic ensemble of the RNA structure of the Leptospira interrogans , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
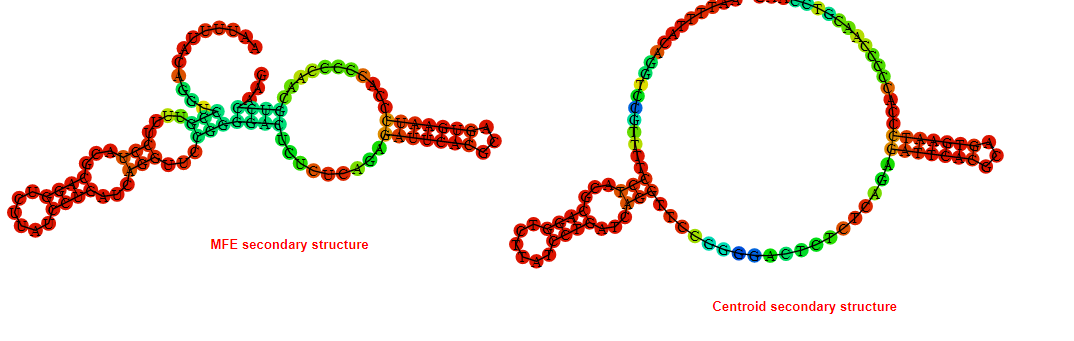
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Leptospira interrogans , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
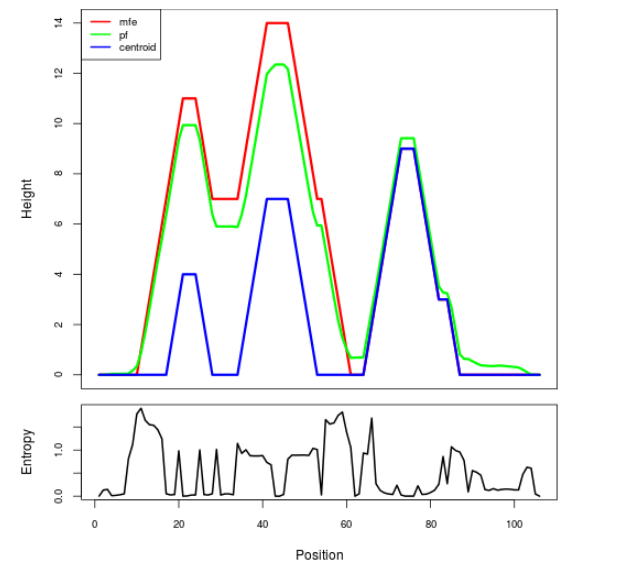
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
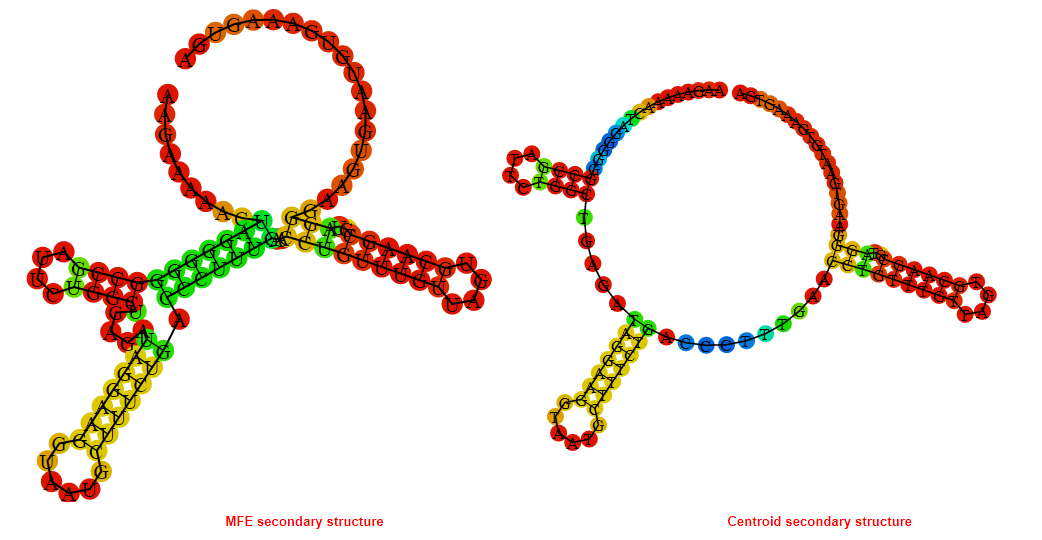
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
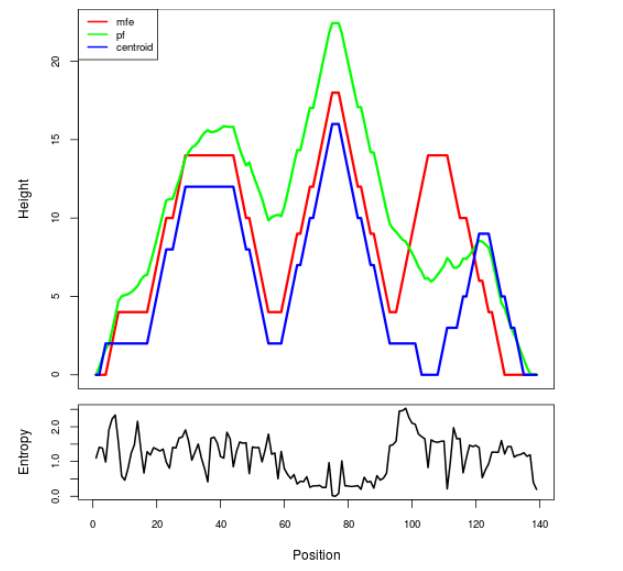
The plot represents the thermodynamic ensemble of the RNA structure of the Mycobacterium leprae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
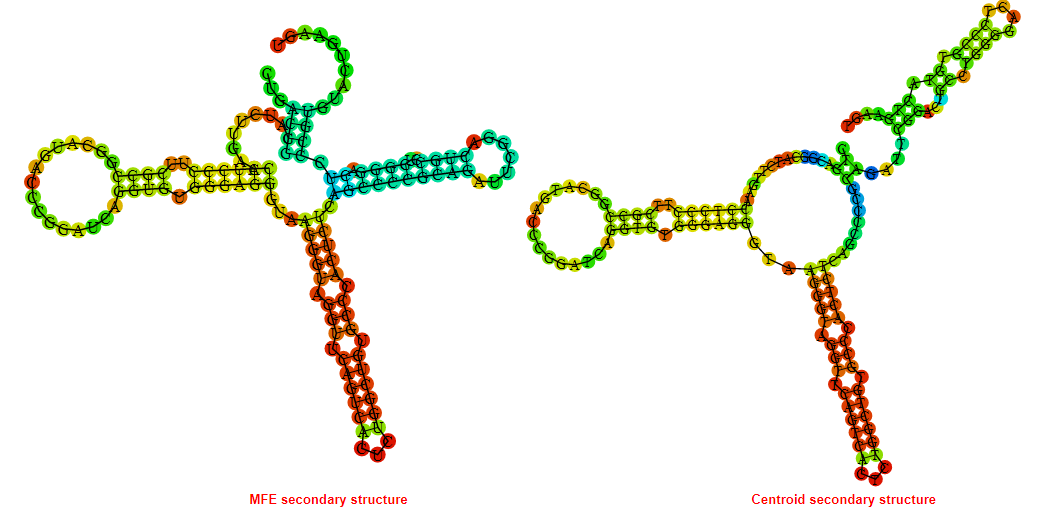
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Mycobacterium leprae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
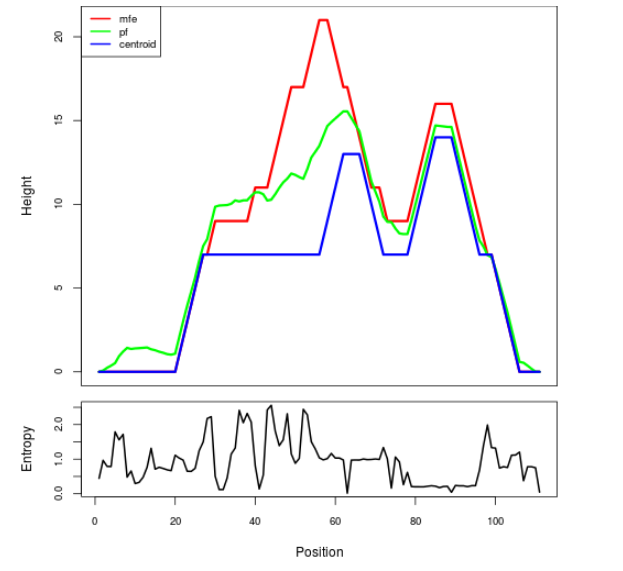
The plot represents the thermodynamic ensemble of the RNA structure of the Mycobacterium tuberculosis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
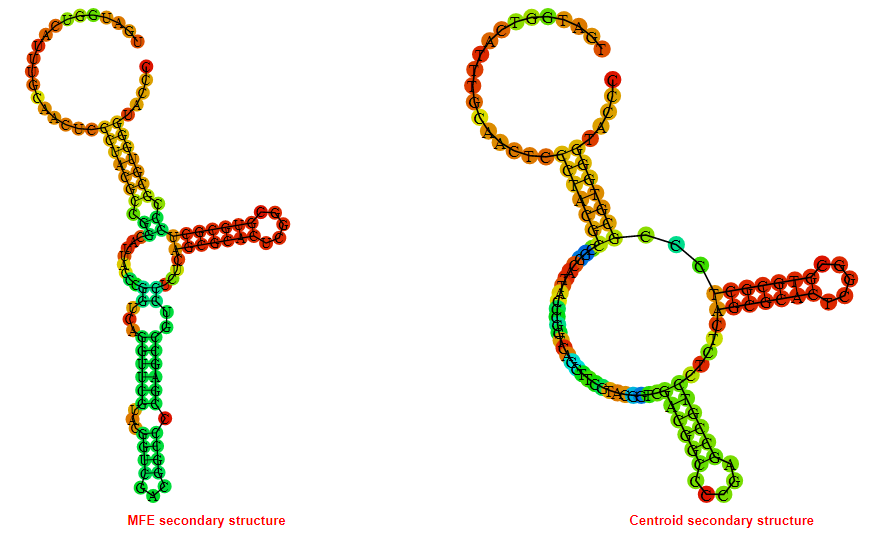
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Mycobacterium tuberculosis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
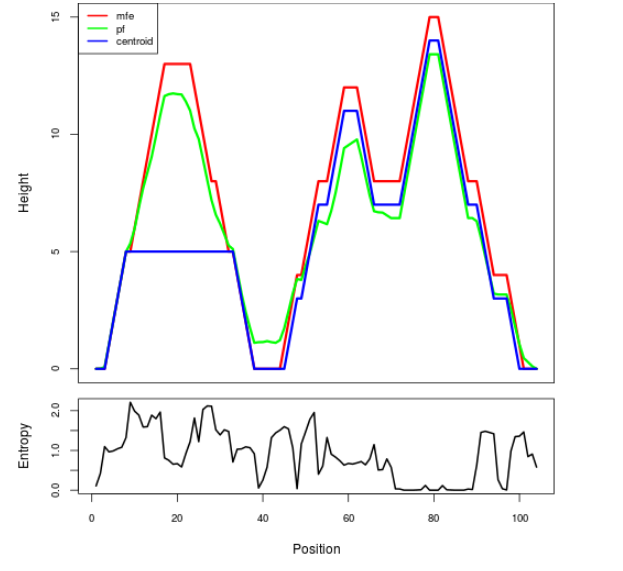
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria gonorrhoeae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria gonorrhoeae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
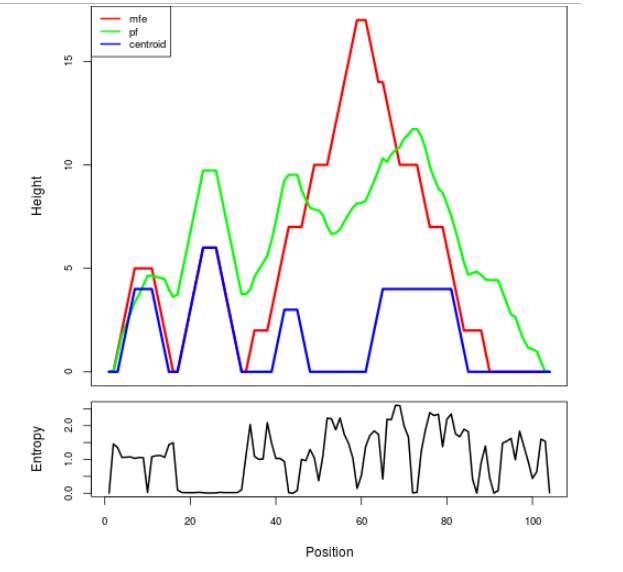
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria meningitidis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
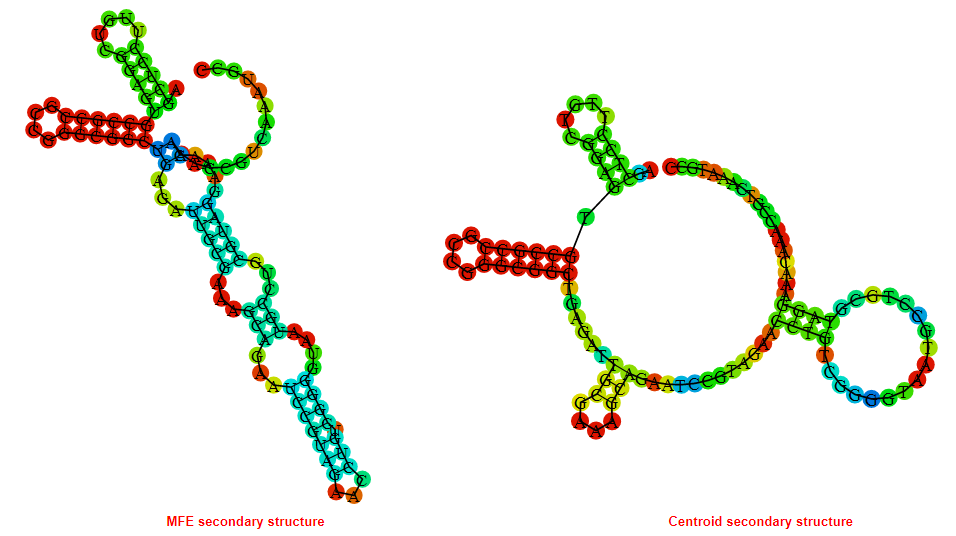
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria meningitidis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
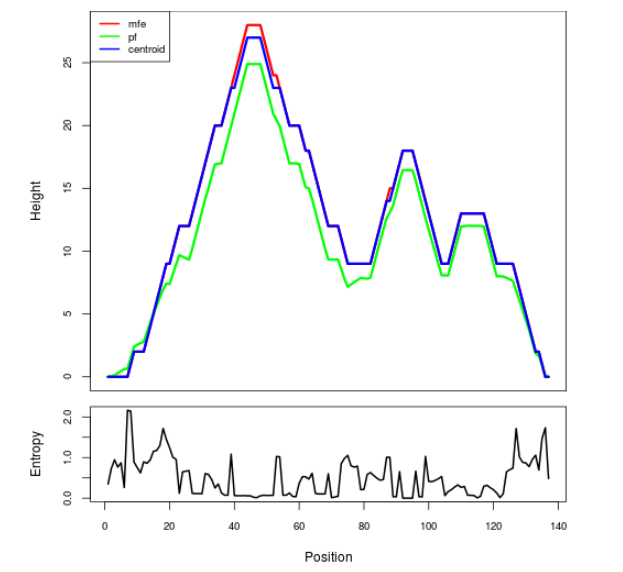
The plot represents the thermodynamic ensemble of the RNA structure of the Pseudomonas aeruginosa , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
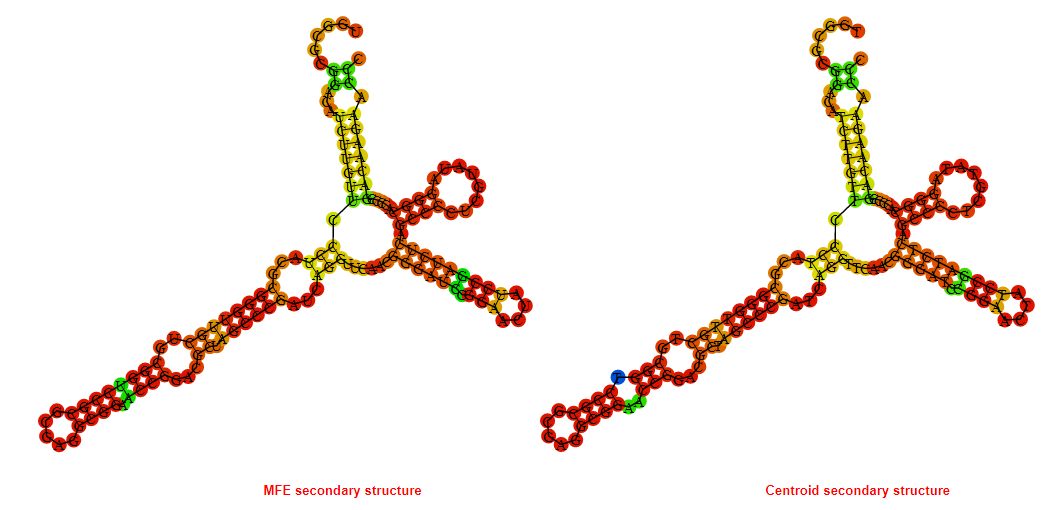
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Pseudomonas aeruginosa , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
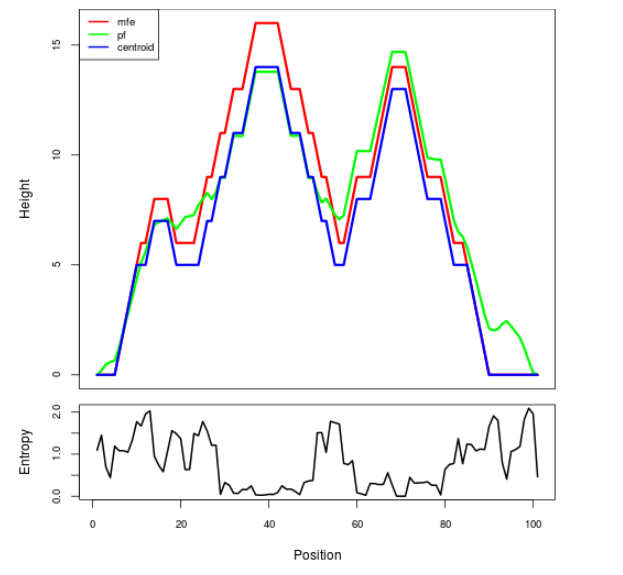
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella enterica , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
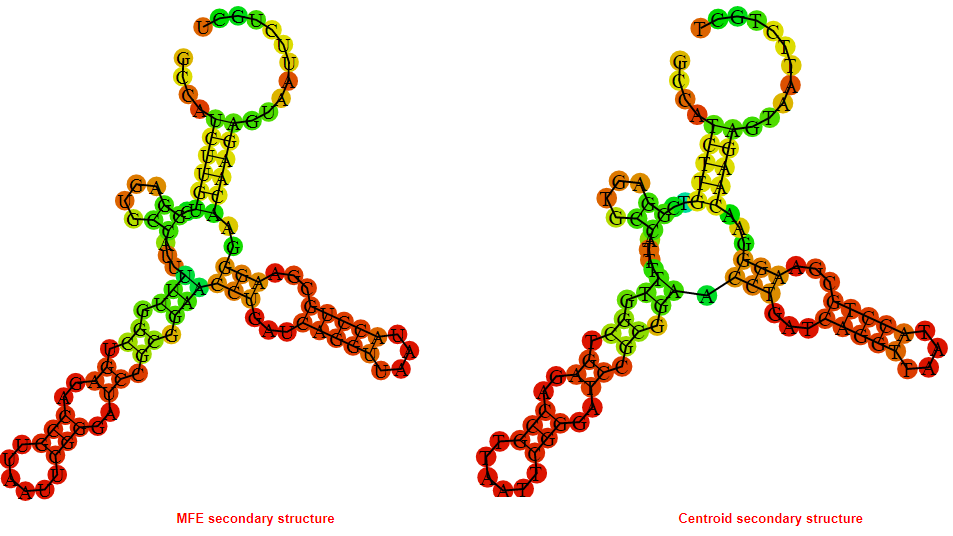
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella enterica , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
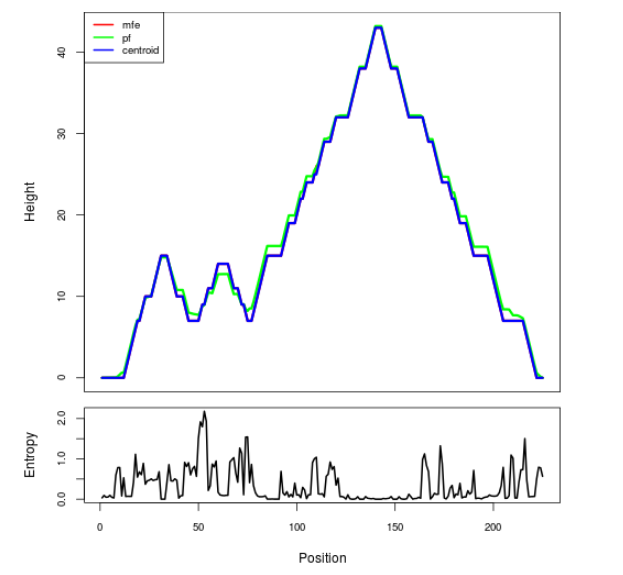
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella Typhimurium , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
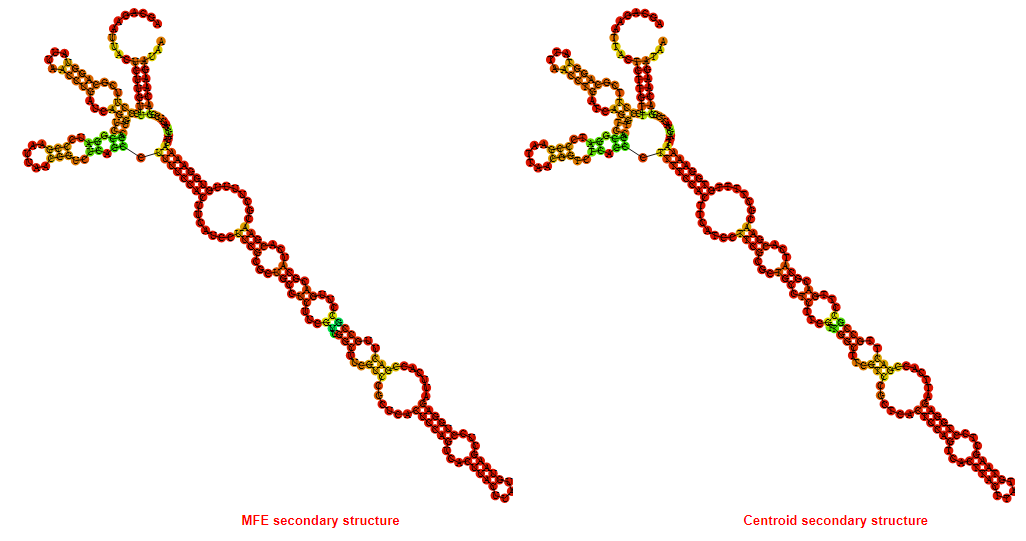
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella Typhimurium , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
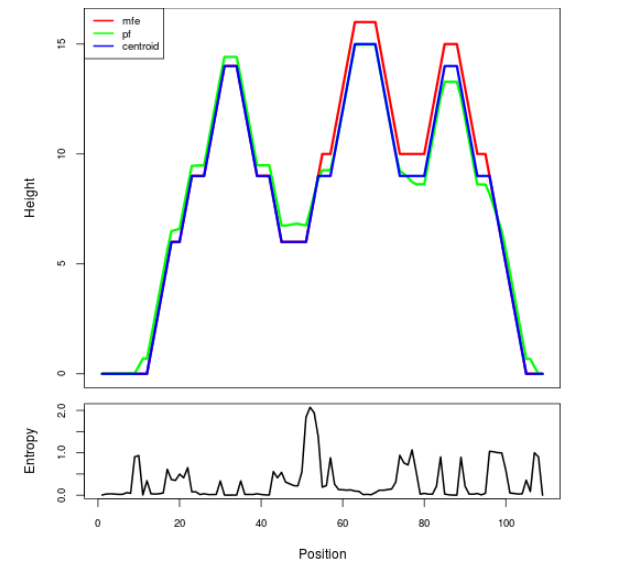
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
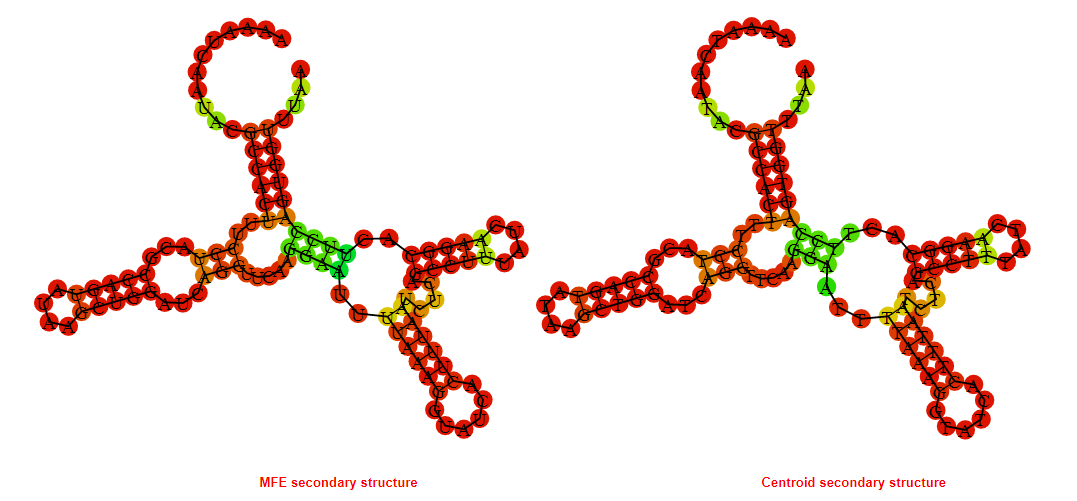
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
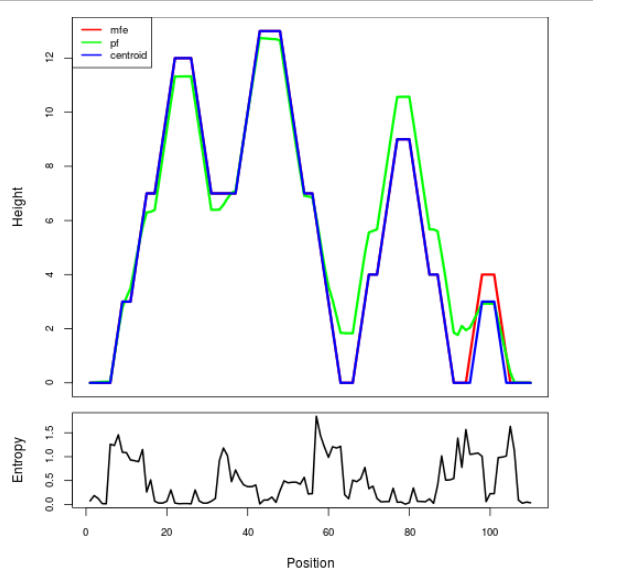
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
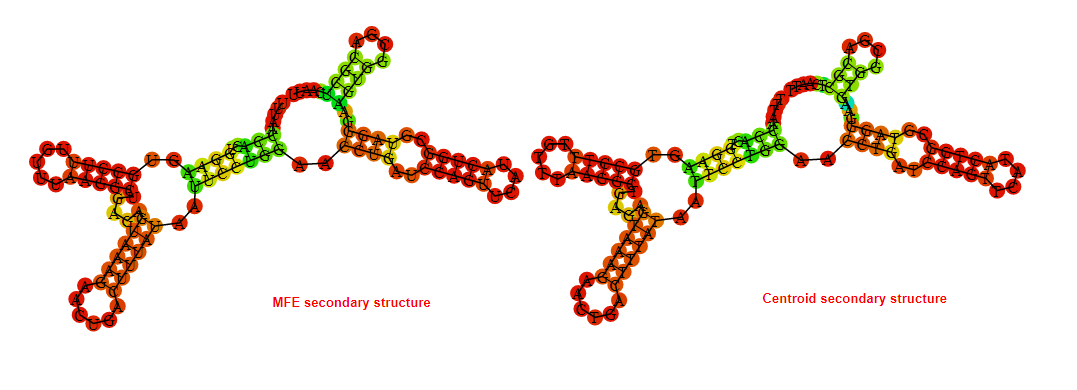
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
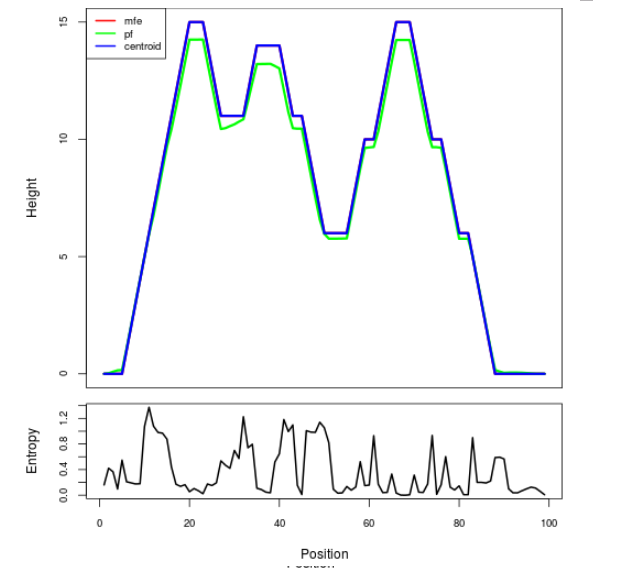
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
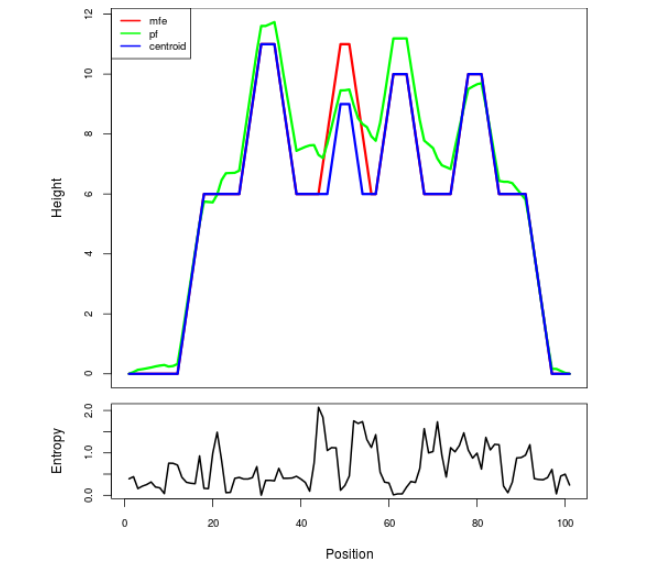
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus mutans , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
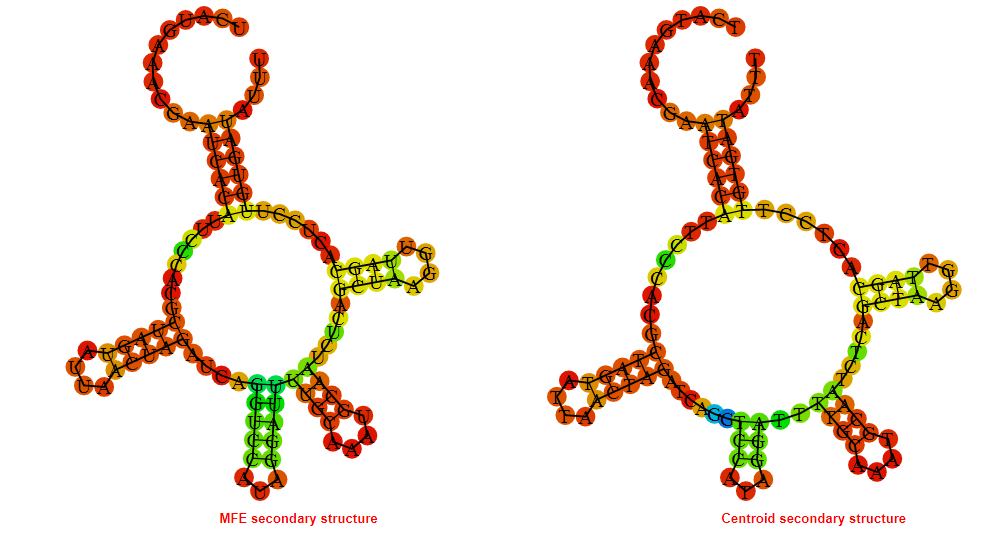
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus mutans , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
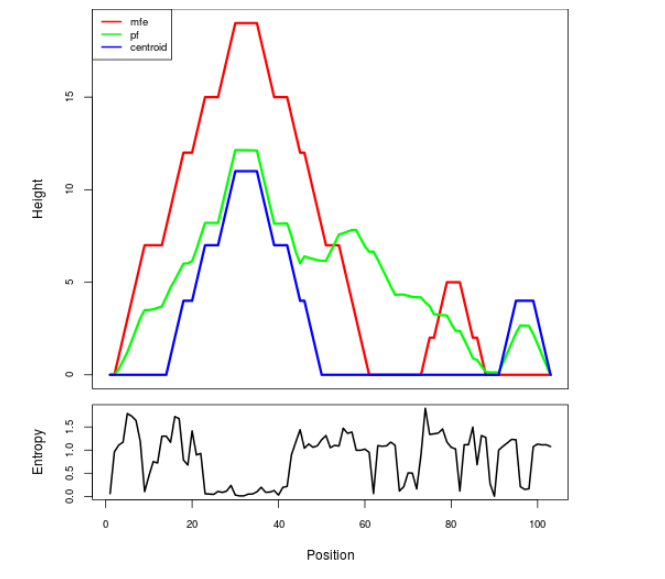
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pneumoniae , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
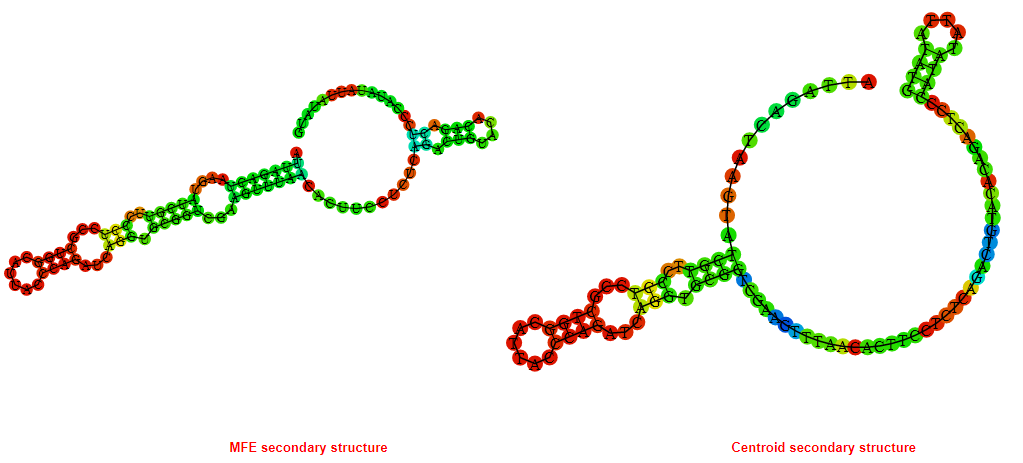
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pneumoniae , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
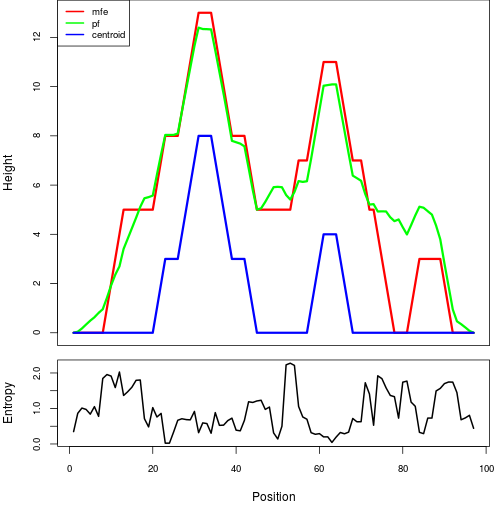
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus pyogenes , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
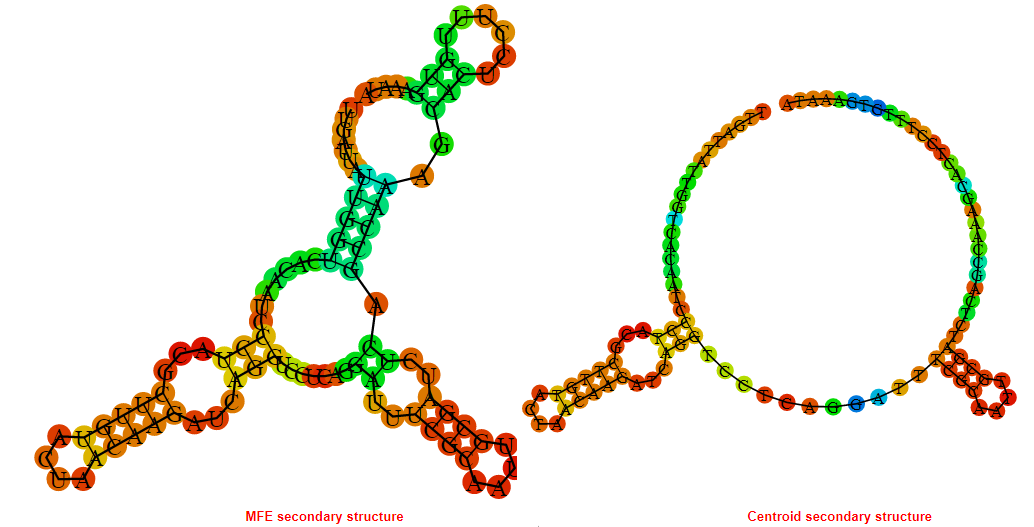
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus pyogenes , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
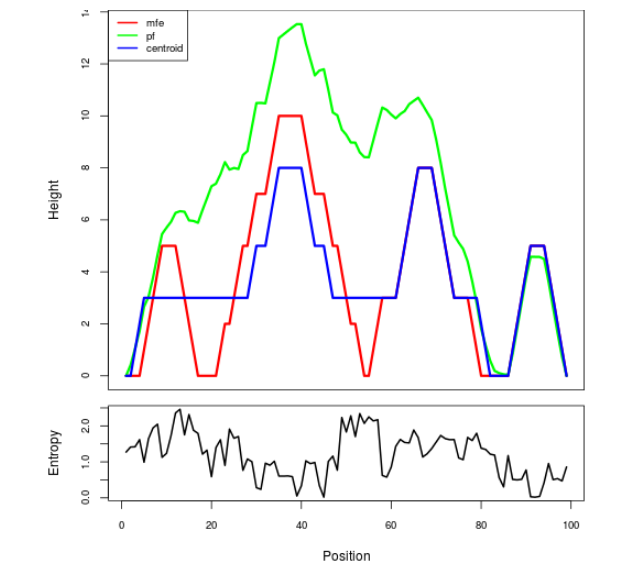
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
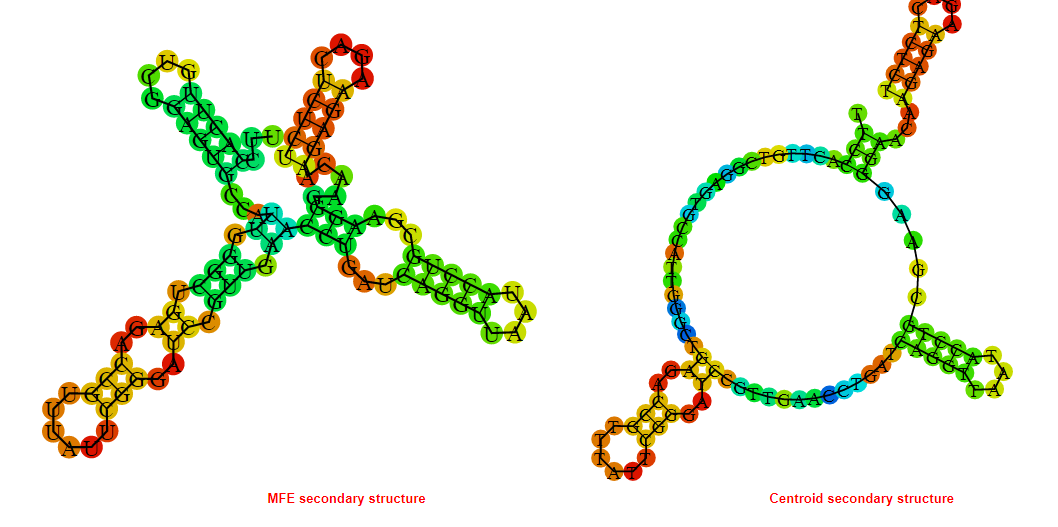
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
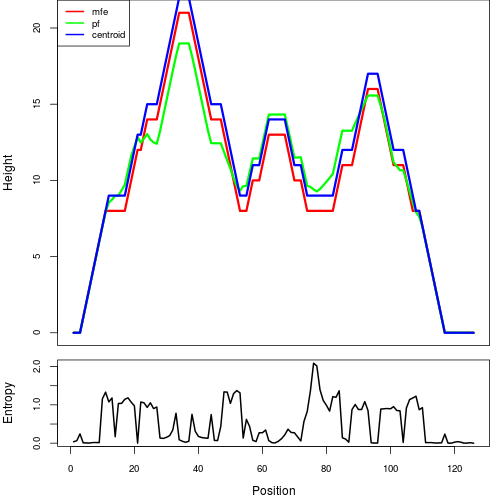
The plot represents the thermodynamic ensemble of the RNA structure of the Yersinia pestis , TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
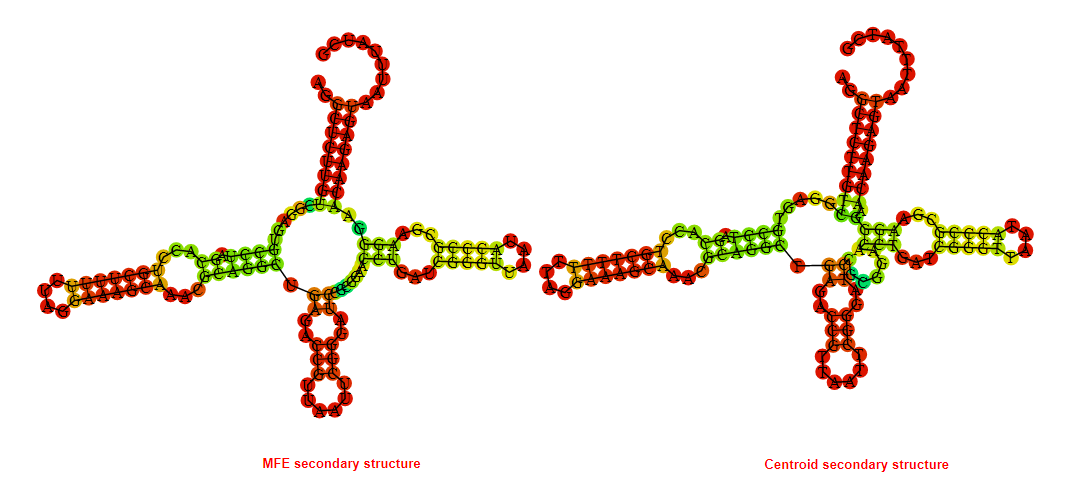
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Yersinia pestis , TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
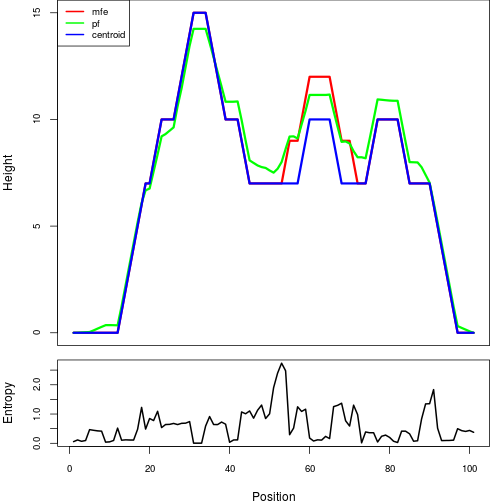
The plot represents the thermodynamic ensemble of the RNA structure of the Escherichia coli, TPP type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
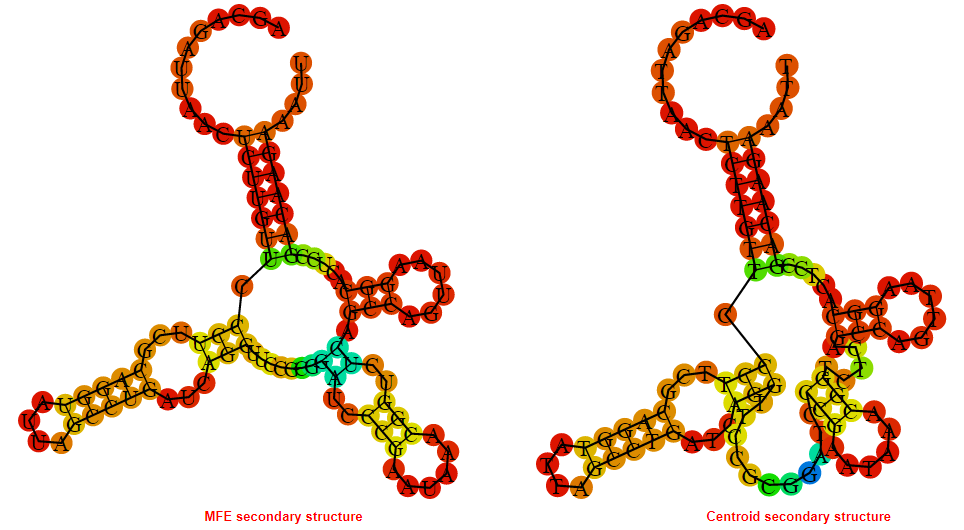
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Escherichia coli, TPP type switch.
Multiple alignment of the TPP type switches
Below is presented the sequences alignment of all the TPP type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the TPP type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the TPP type switches involved in.

Multi_resistance strains for the TPP type switches
The table displays known human pathogenic bacteria bearing the TPP type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
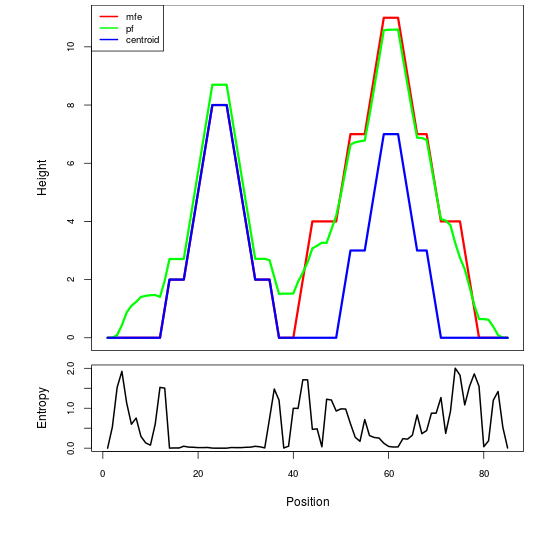
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
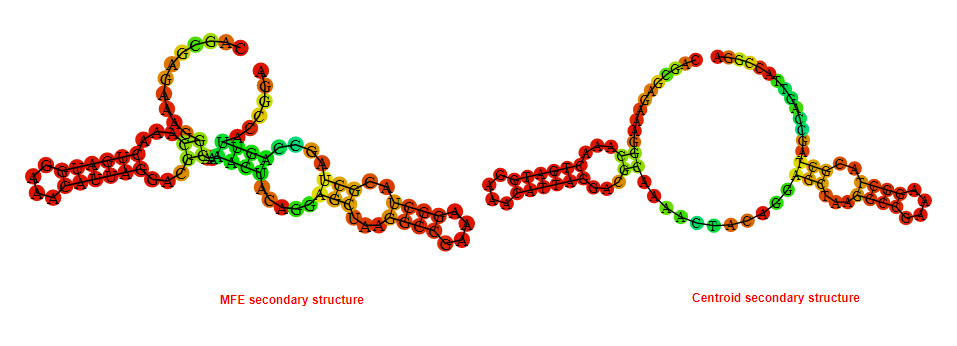
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.

Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
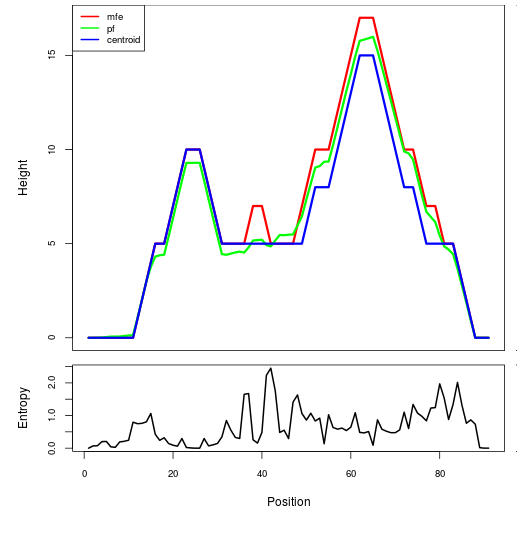
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
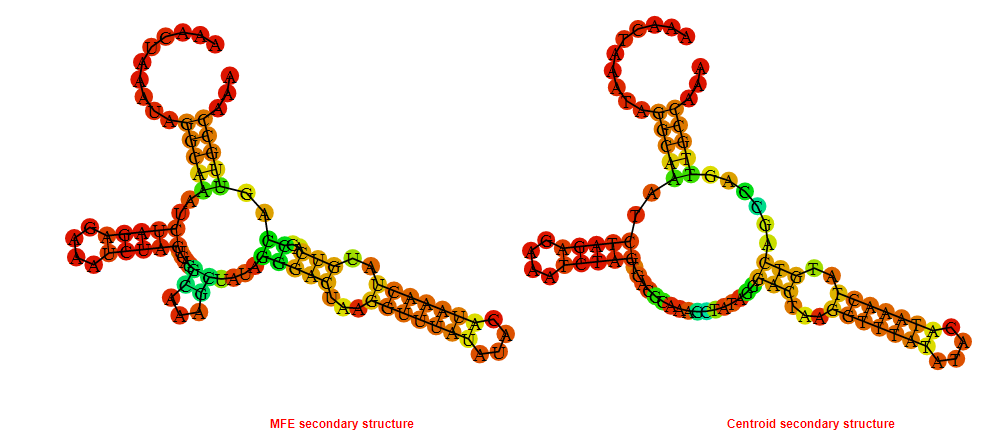
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.

Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
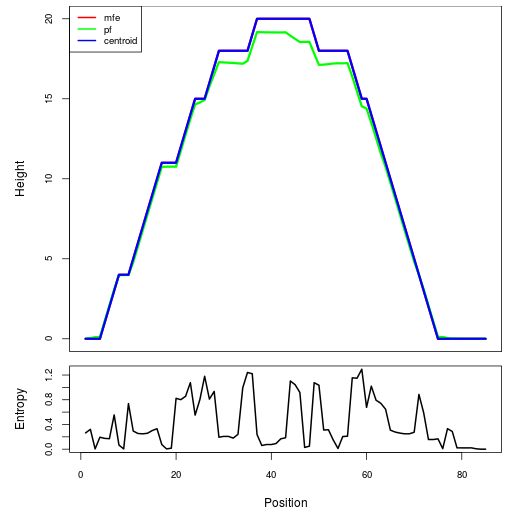
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum, Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum, Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.
Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
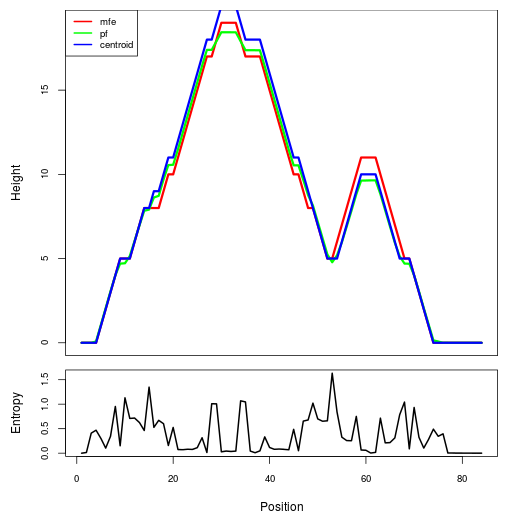
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens, Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
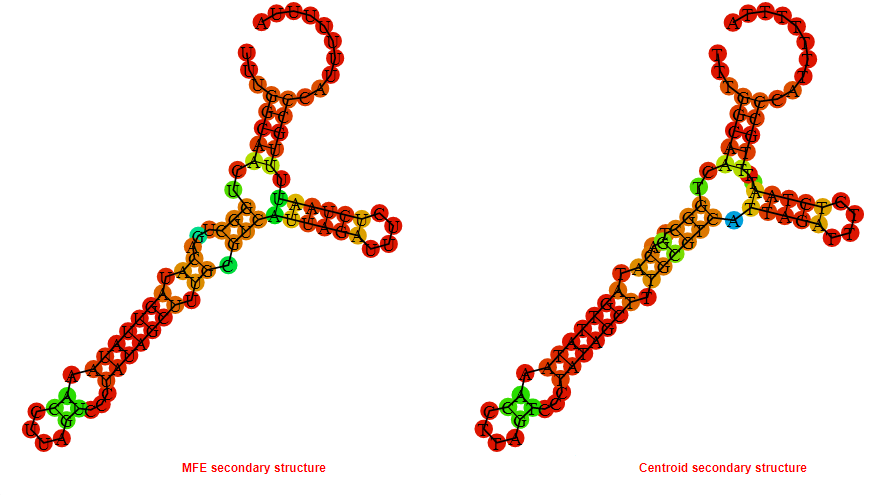
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens, Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.

Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
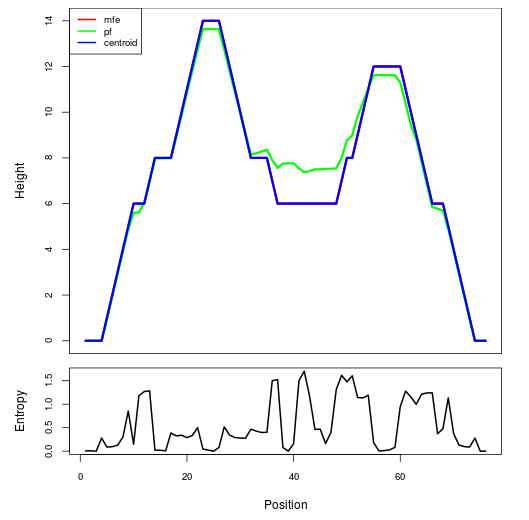
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani, Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
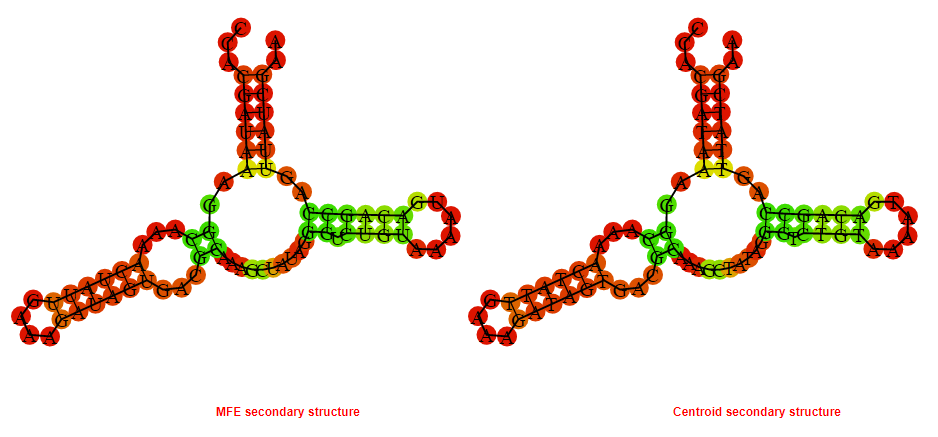
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani, Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.

Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
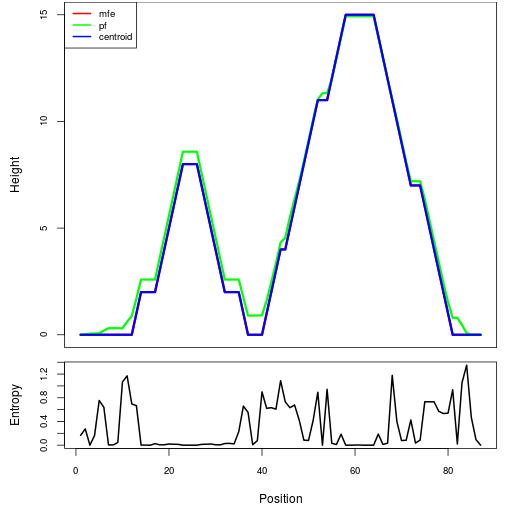
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae, Cyclic di-GMP I type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
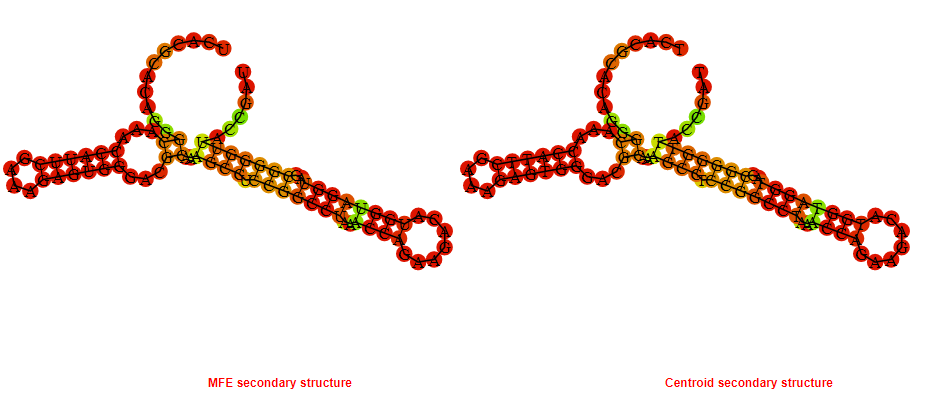
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae, Cyclic di-GMP I type switch.
Multiple alignment of the Cyclic di-GMP I type switches
Below is presented the sequences alignment of all the Cyclic di-GMP I type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP I type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP I type switches involved in.

Multi_resistance strains for the Cyclic di-GMP I type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP I type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
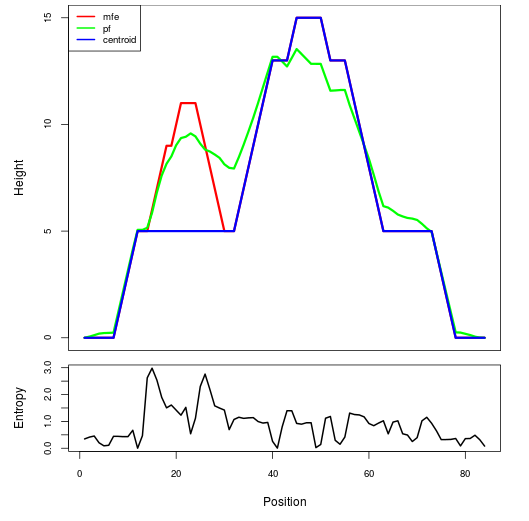
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum, Cyclic di-GMP II type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
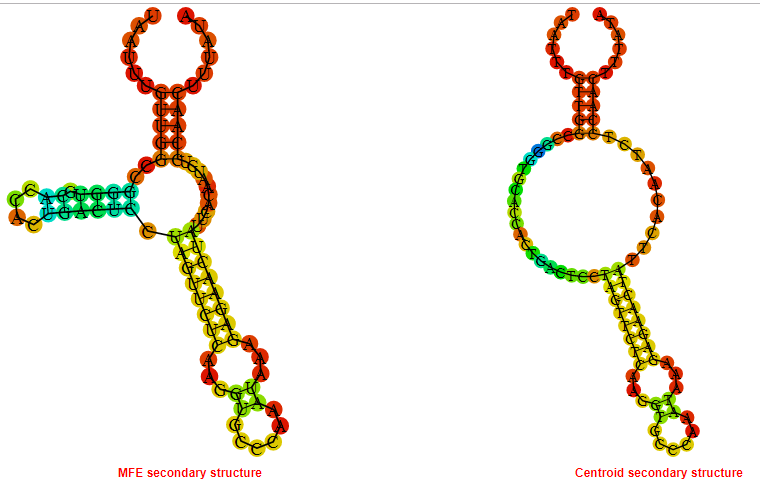
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum, Cyclic di-GMP II type switch.
Multiple alignment of the Cyclic di-GMP II type switches
Below is presented the sequences alignment of all the Cyclic di-GMP II type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP II type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP II type switches involved in.
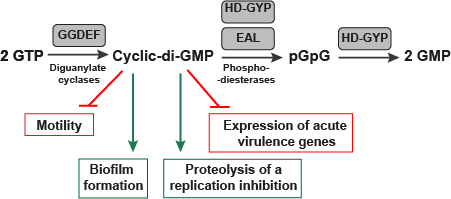
Multi_resistance strains for the Cyclic di-GMP II type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP II type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
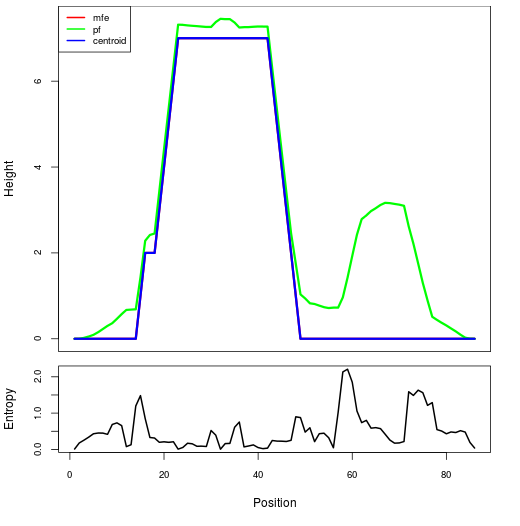
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile, Cyclic di-GMP II type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
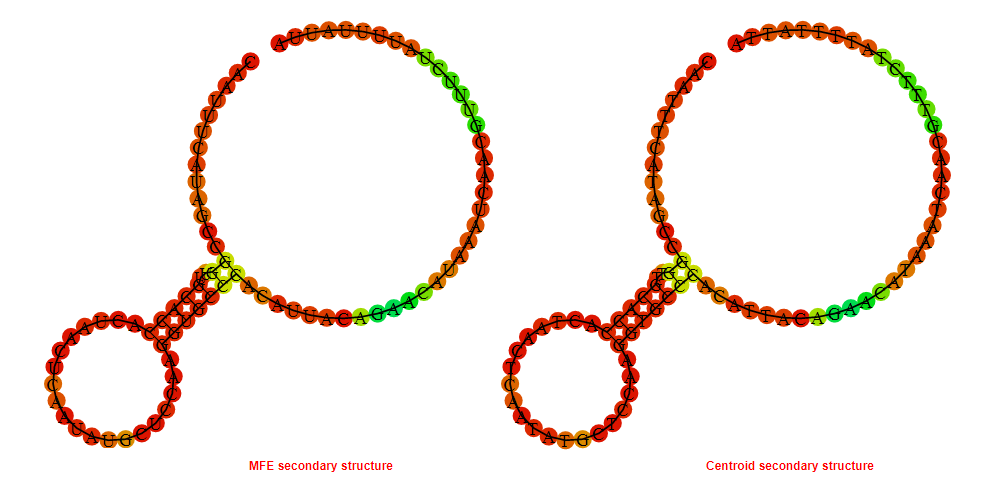
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile, Cyclic di-GMP II type switch.
Multiple alignment of the Cyclic di-GMP II type switches
Below is presented the sequences alignment of all the Cyclic di-GMP II type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP II type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP II type switches involved in.

Multi_resistance strains for the Cyclic di-GMP II type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP II type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
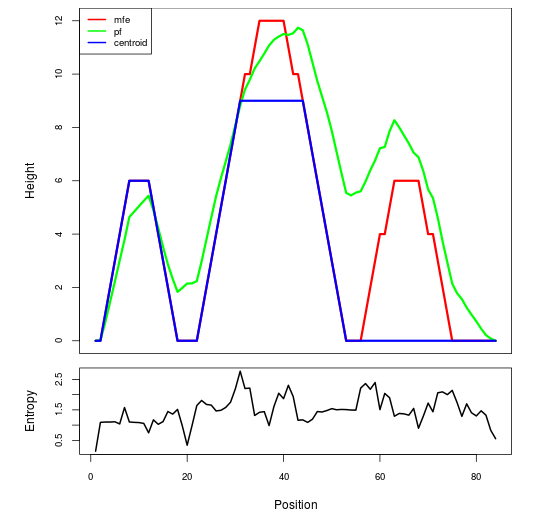
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens, Cyclic di-GMP II type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
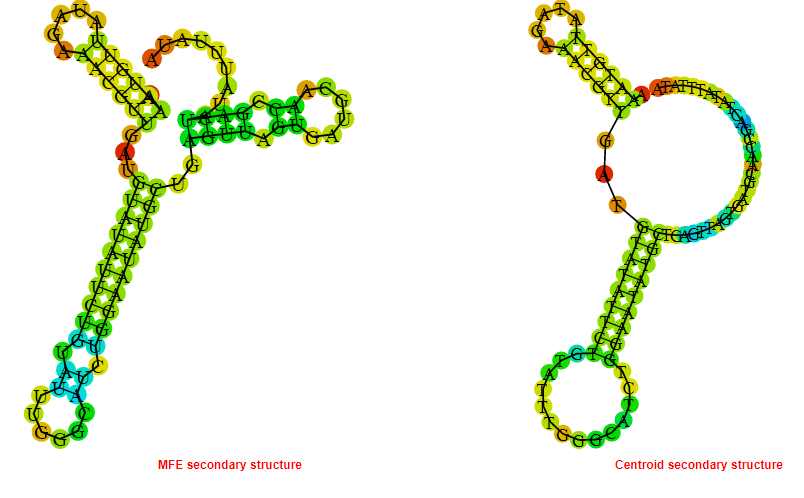
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens, Cyclic di-GMP II type switch.
Multiple alignment of the Cyclic di-GMP II type switches
Below is presented the sequences alignment of all the Cyclic di-GMP II type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP II type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP II type switches involved in.

Multi_resistance strains for the Cyclic di-GMP II type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP II type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
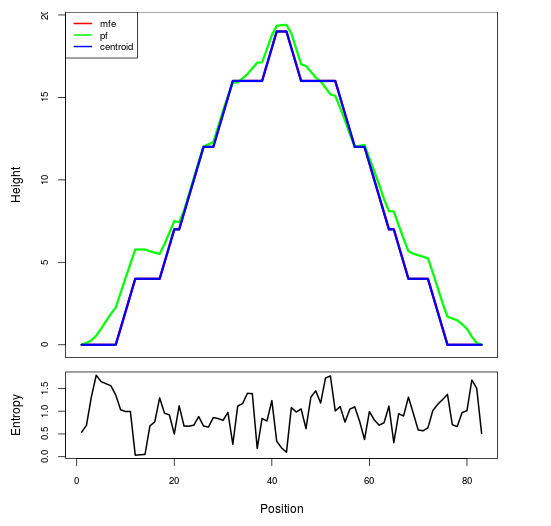
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani, Cyclic di-GMP II type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
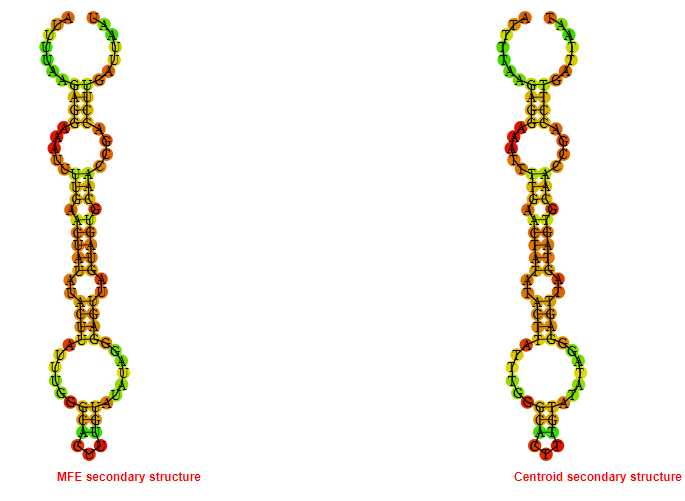
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani, Cyclic di-GMP II type switch.
Multiple alignment of the Cyclic di-GMP II type switches
Below is presented the sequences alignment of all the Cyclic di-GMP II type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Cyclic di-GMP II type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Cyclic di-GMP II type switches involved in.

Multi_resistance strains for the Cyclic di-GMP II type switches
The table displays known human pathogenic bacteria bearing the Cyclic di-GMP II type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
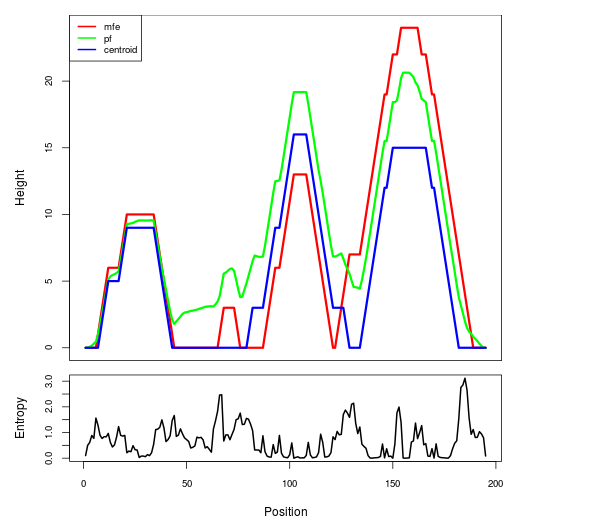
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens, MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
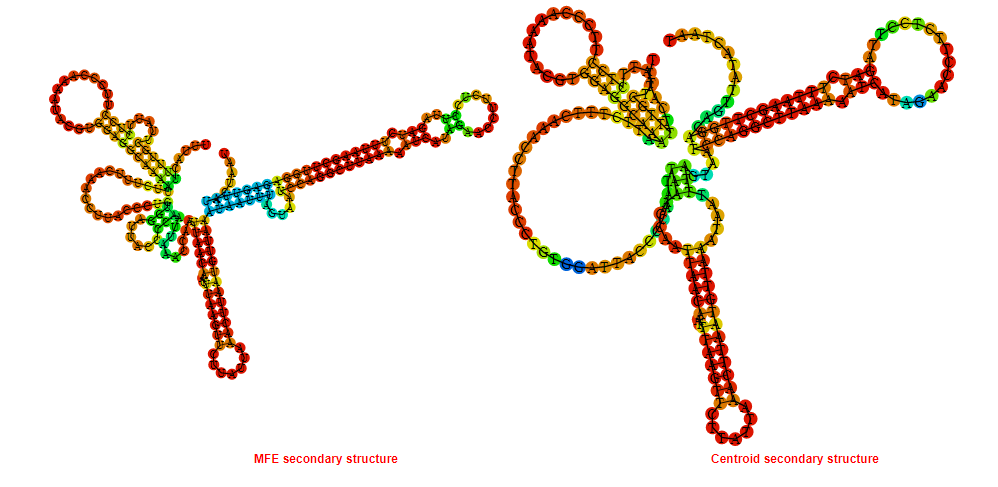
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens, MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
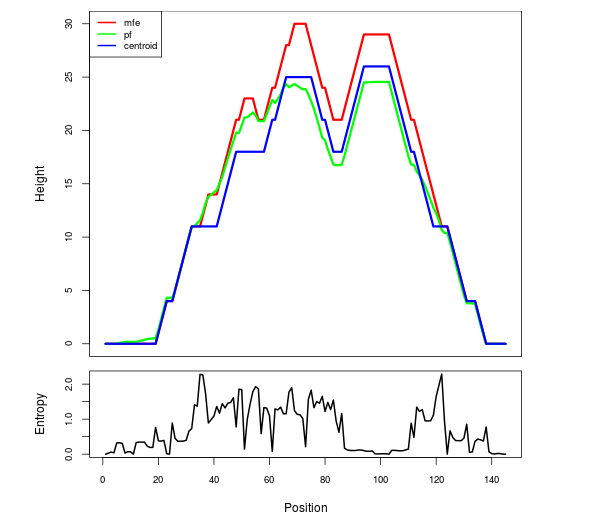
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium coli, MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
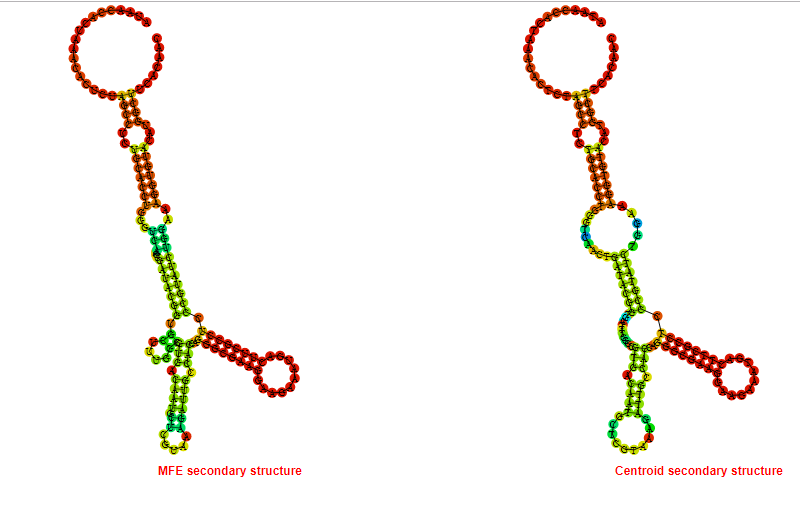
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium coli, MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
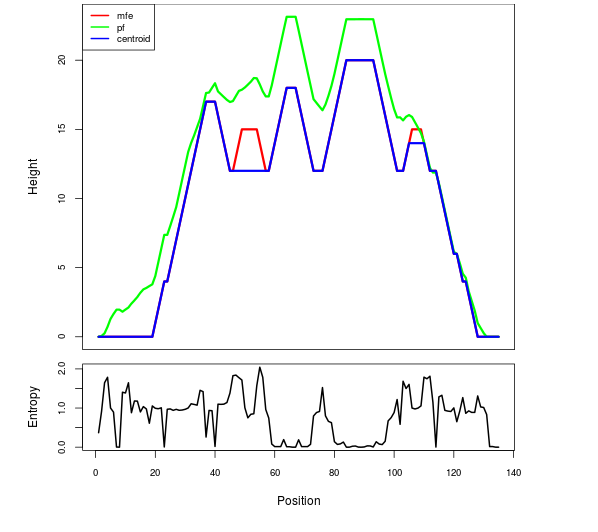
The plot represents the thermodynamic ensemble of the RNA structure of the Enterobacter sp. , MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
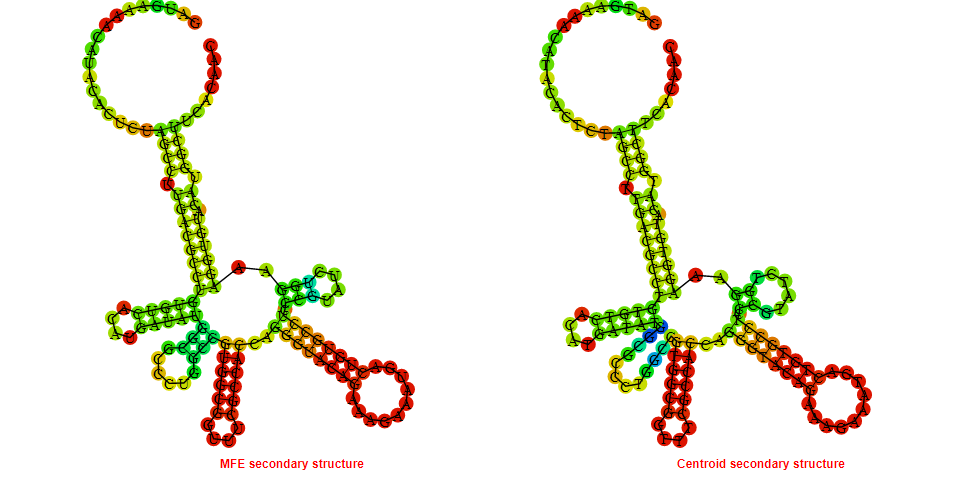
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterobacter sp. , MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
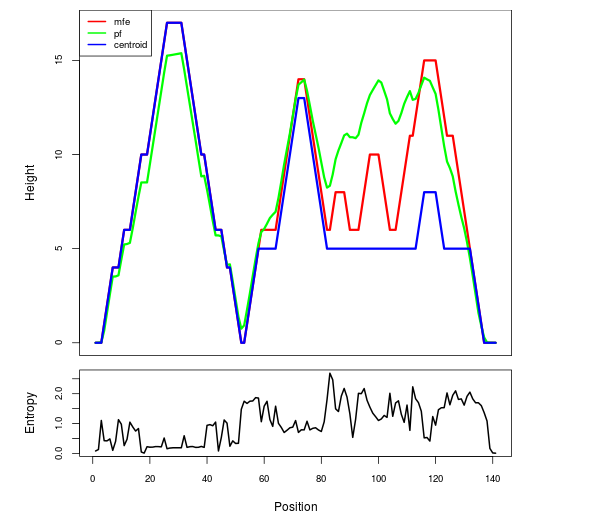
The plot represents the thermodynamic ensemble of the RNA structure of the Haemophilus influenzae, MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
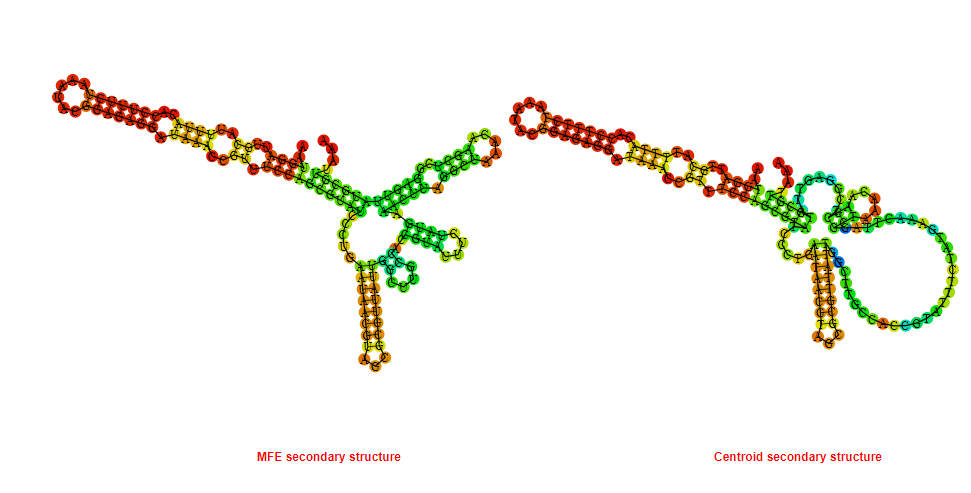
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Haemophilus influenzae, MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.
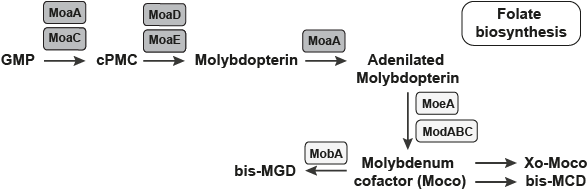
Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
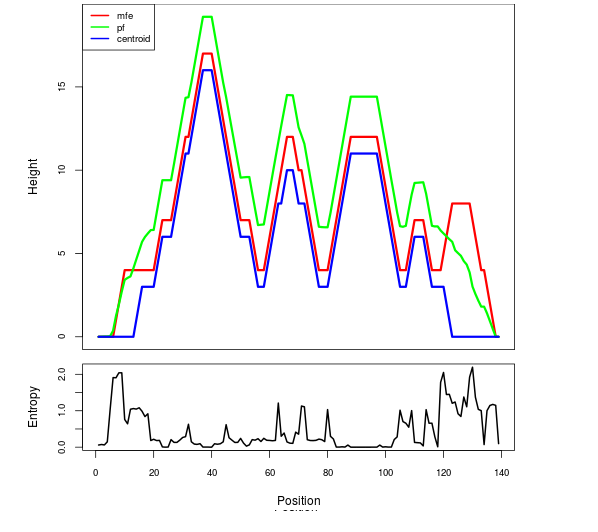
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae, MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
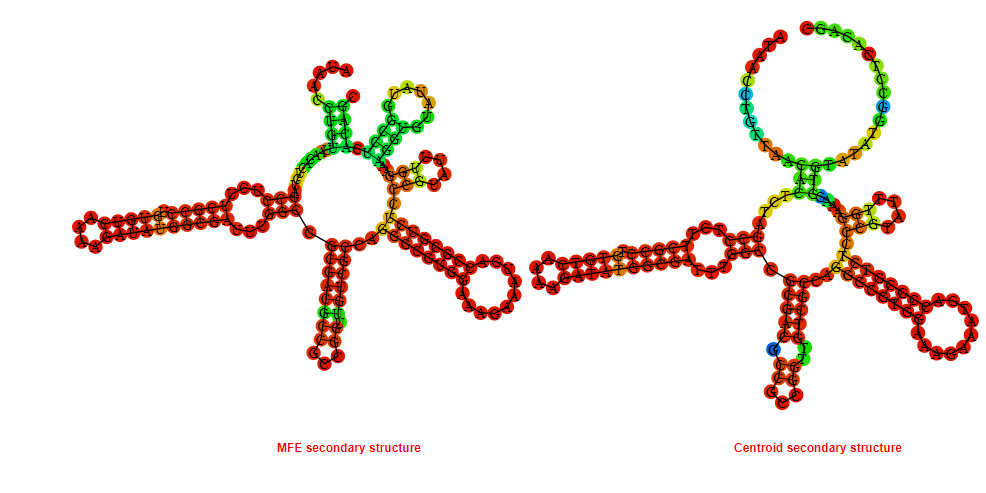
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae, MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
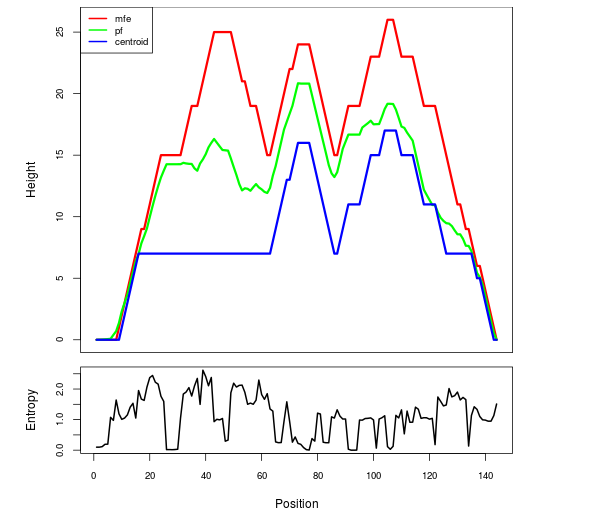
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella enterica, MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
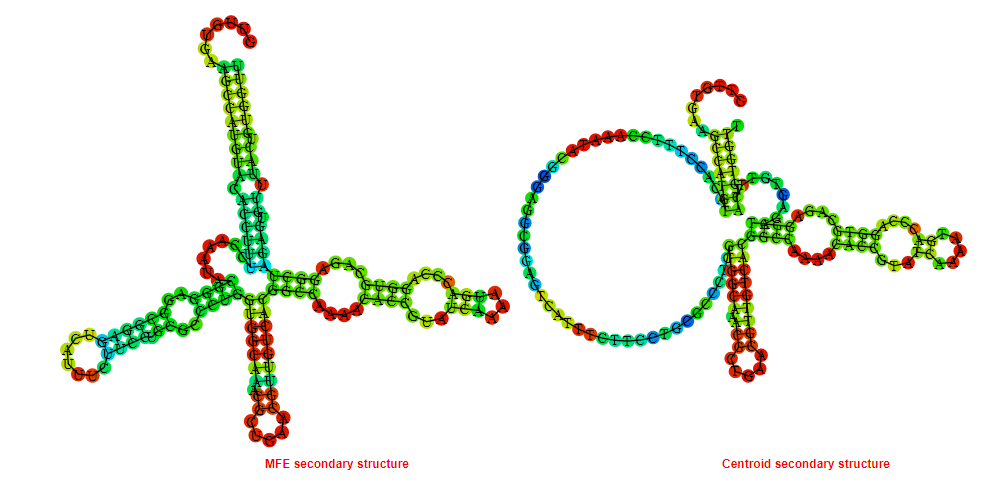
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella enterica, MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
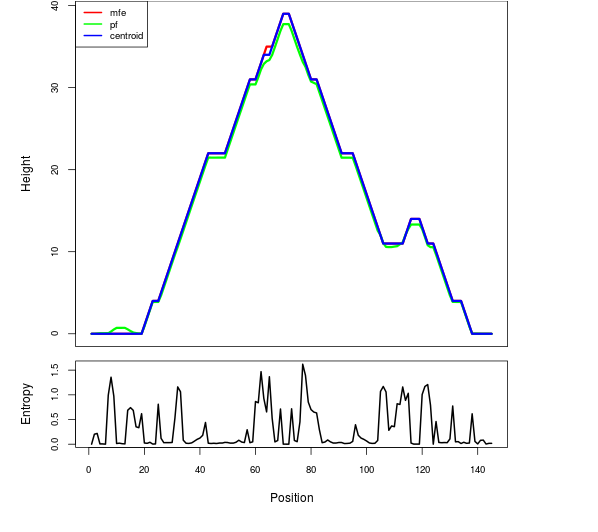
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella typhi , MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
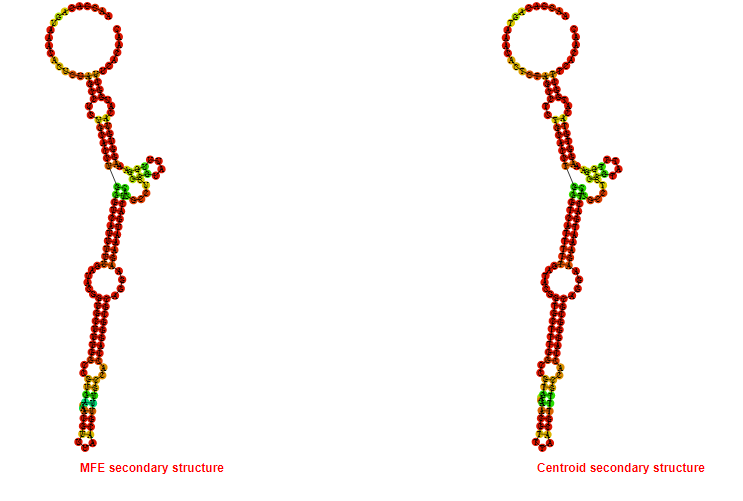
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella typhi , MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
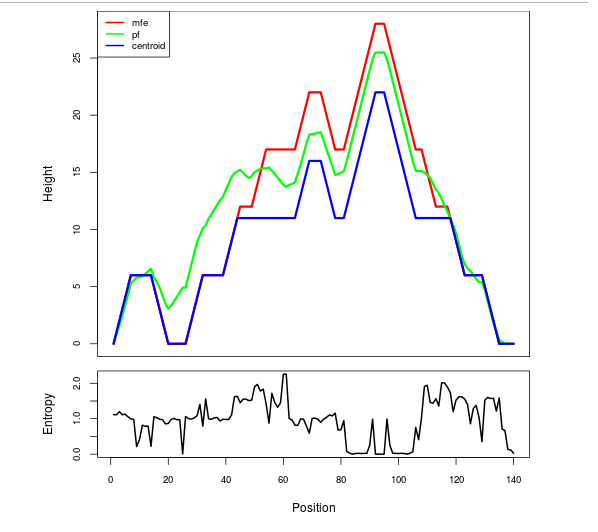
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae , MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
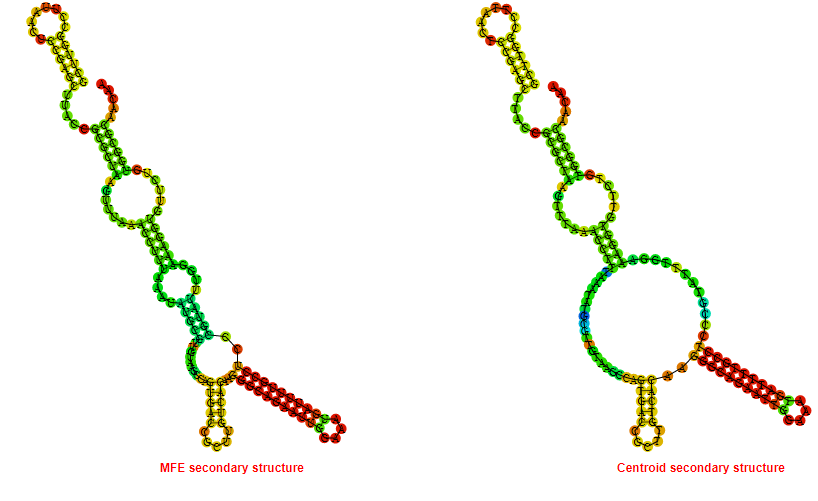
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae , MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
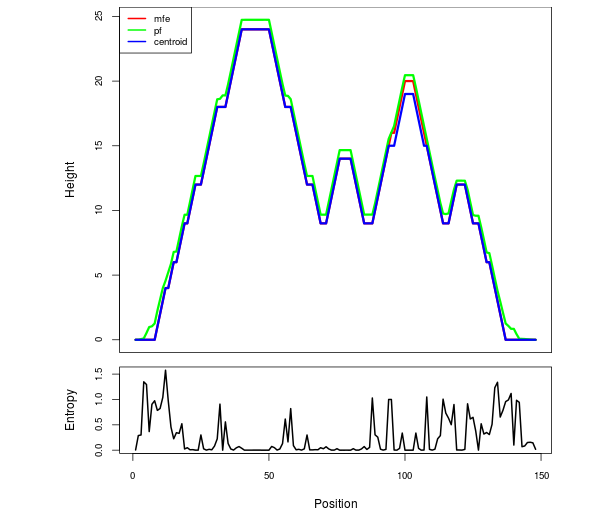
The plot represents the thermodynamic ensemble of the RNA structure of the Yersinia pestis , MOCO type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
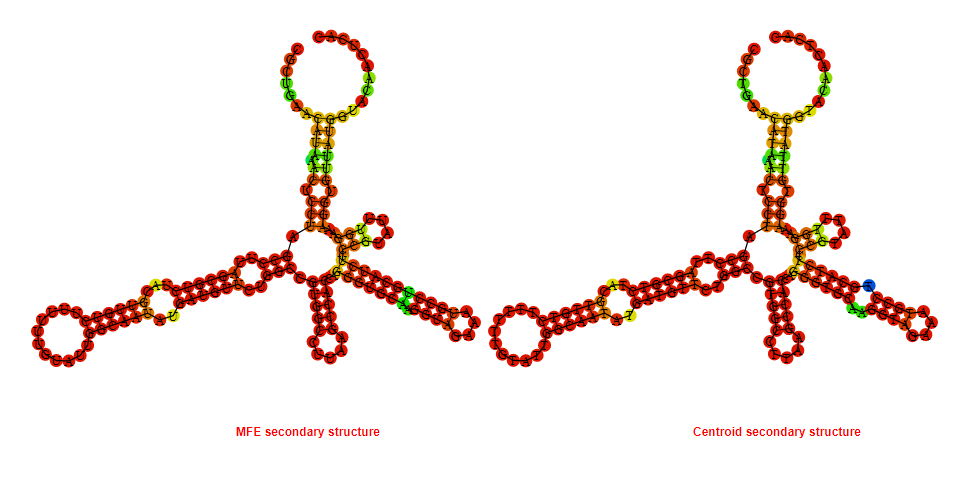
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Yersinia pestis , MOCO type switch.
Multiple alignment of the MOCO type switches
Below is presented the sequences alignment of all the MOCO type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the MOCO type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the MOCO type switches involved in.

Multi_resistance strains for the MOCO type switches
The table displays known human pathogenic bacteria bearing the MOCO type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
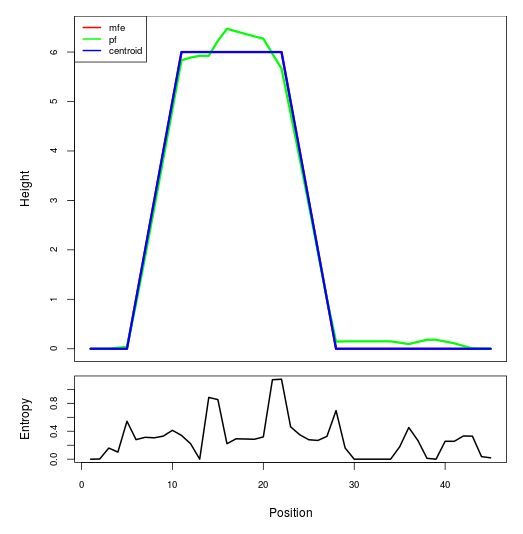
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
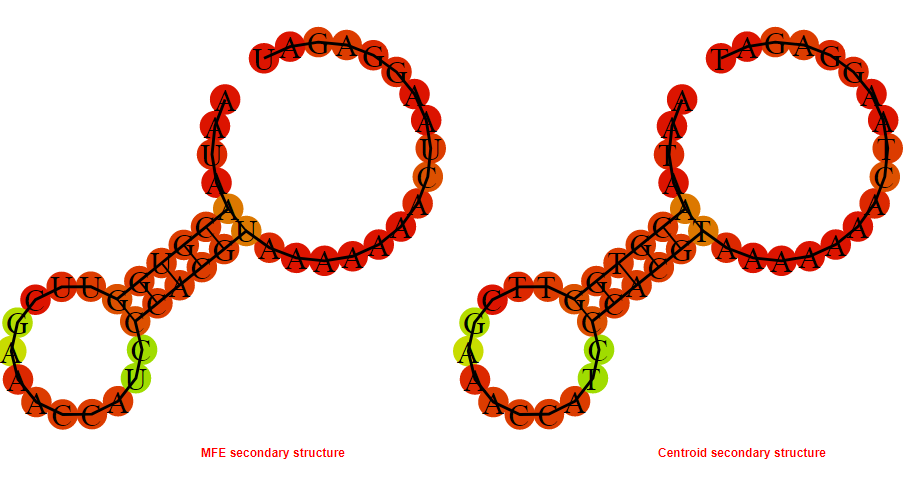
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.
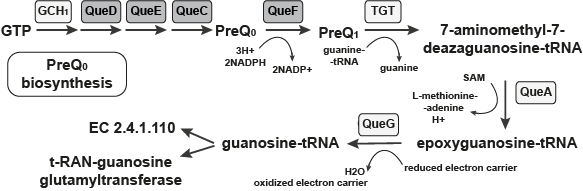
Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
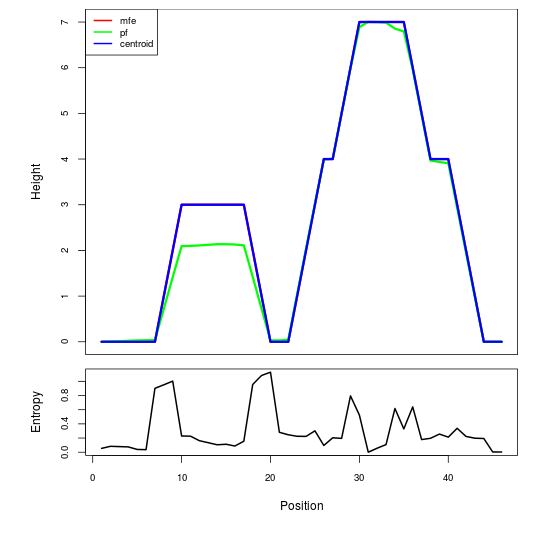
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
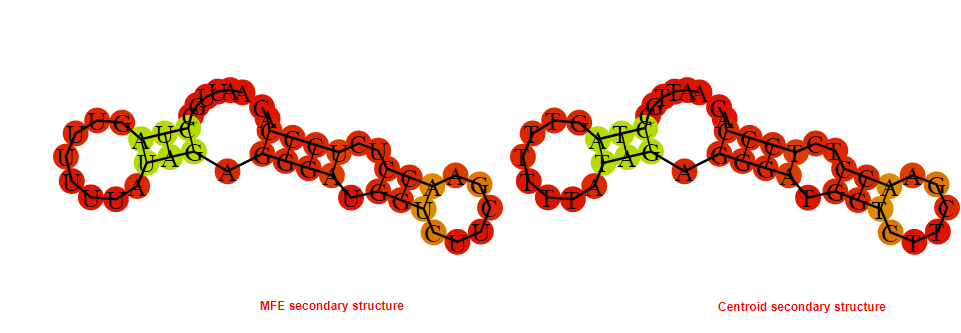
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
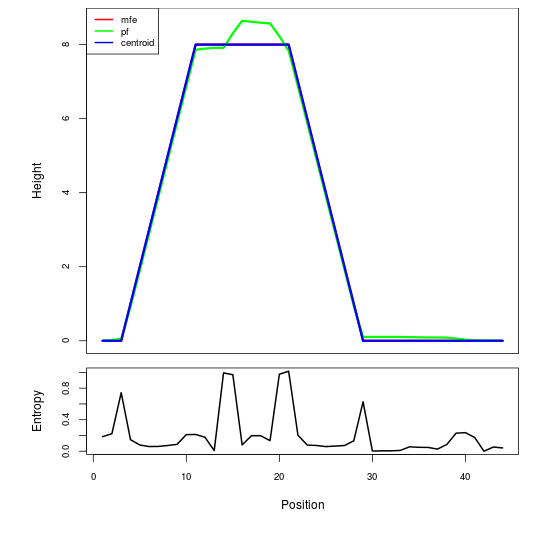
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
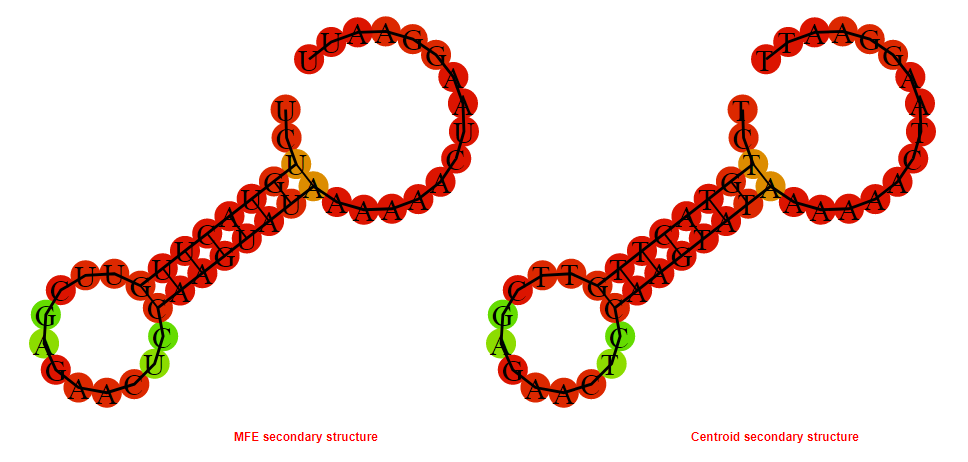
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
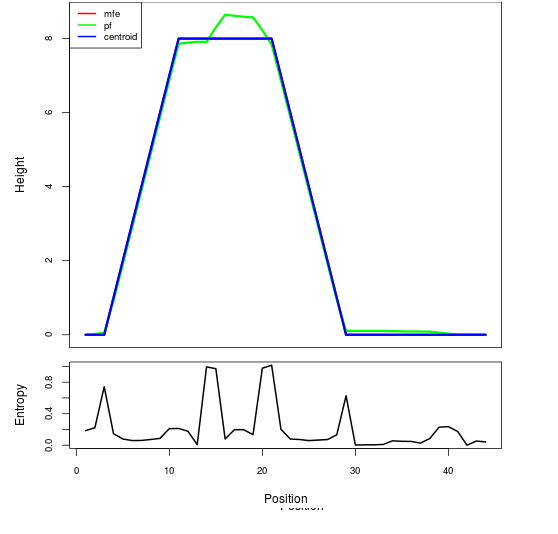
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
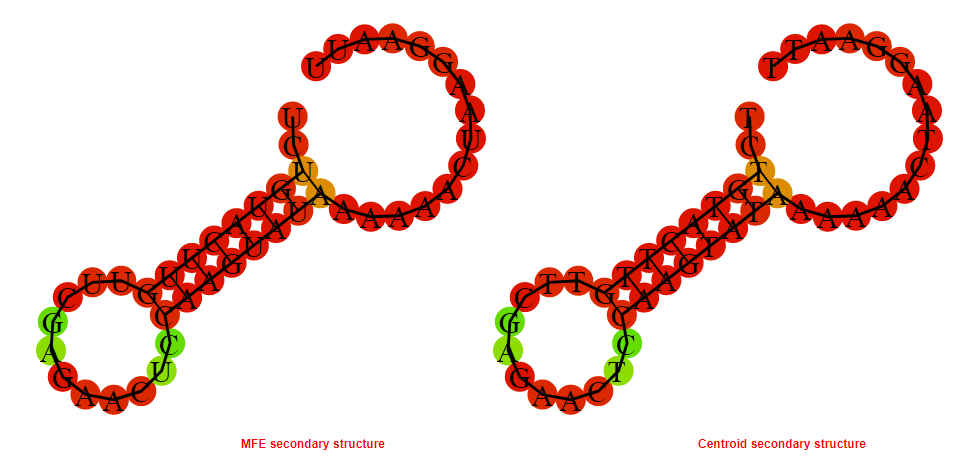
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
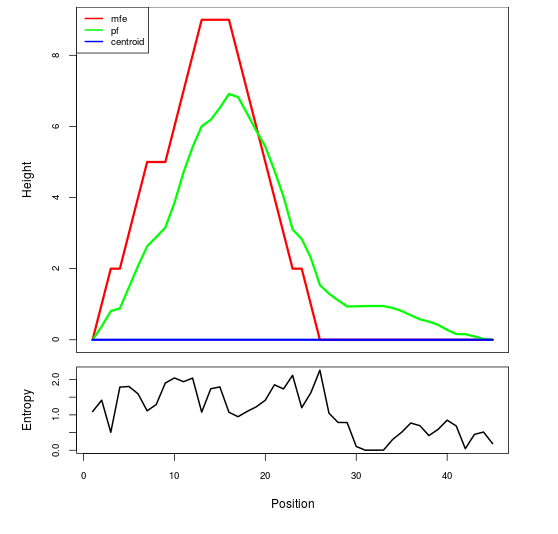
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
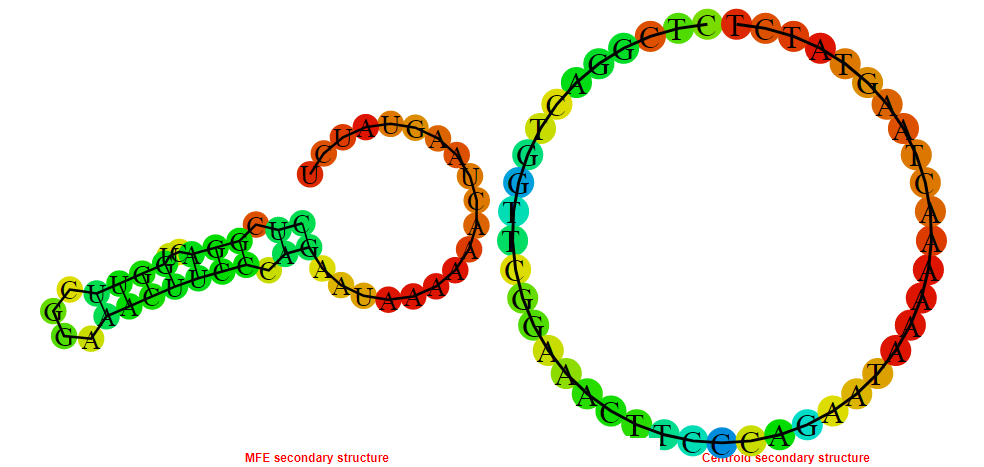
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
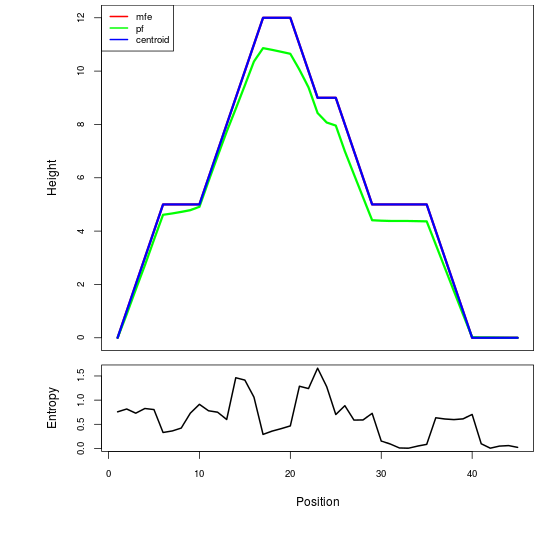
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
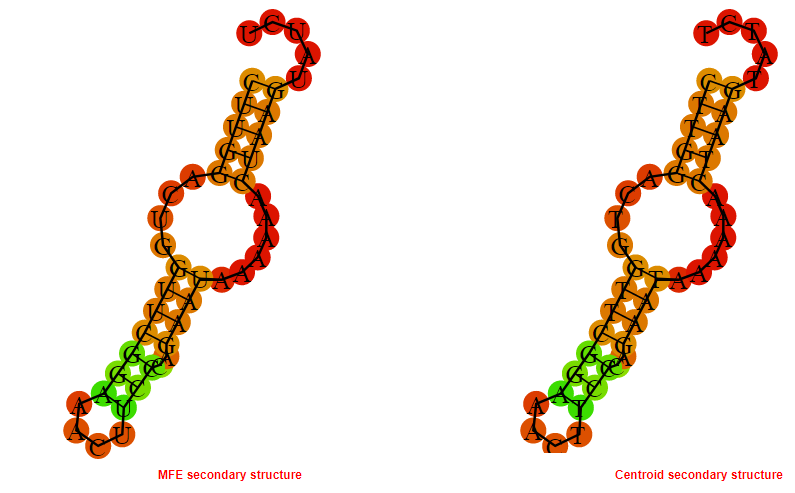
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
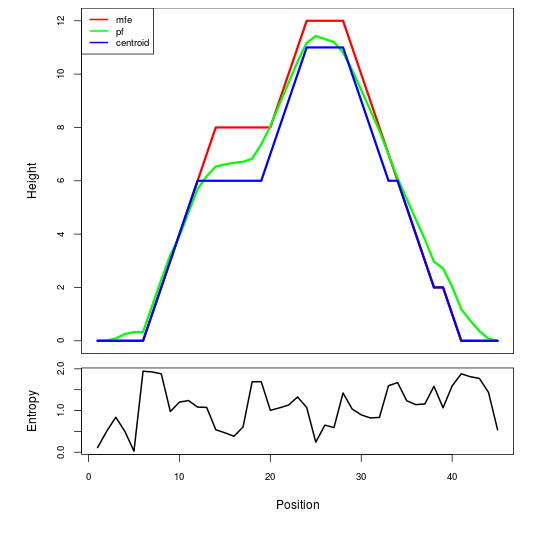
The plot represents the thermodynamic ensemble of the RNA structure of the Haemophilus influenzae , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
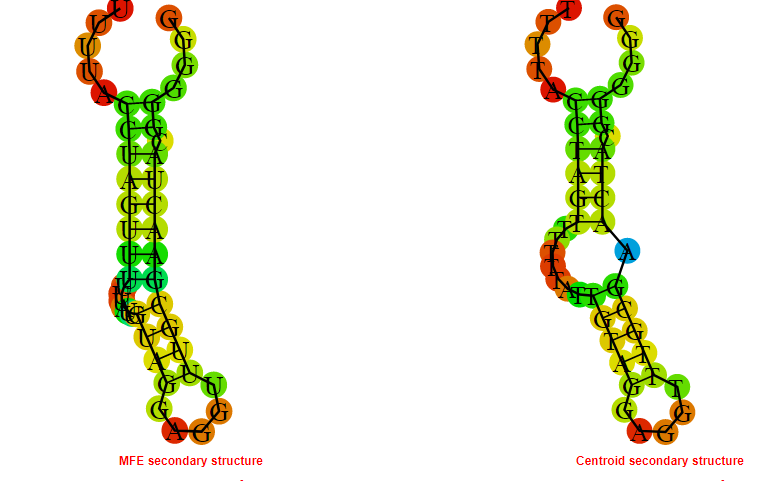
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Haemophilus influenzae , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
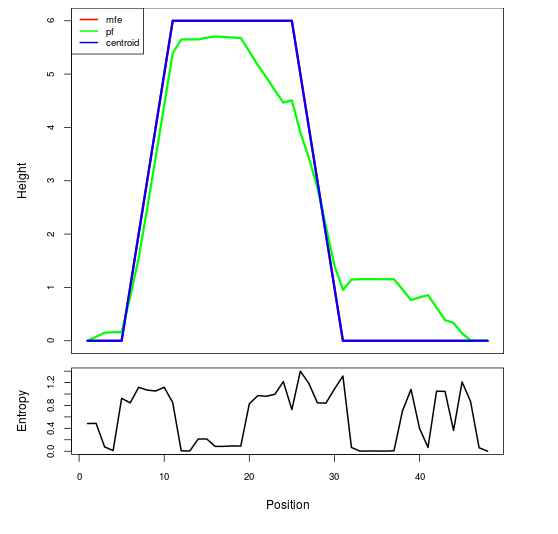
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
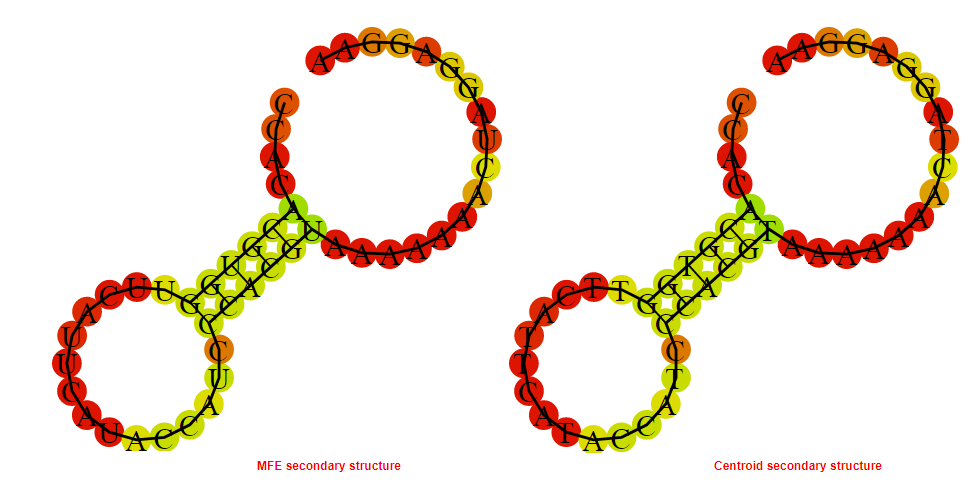
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
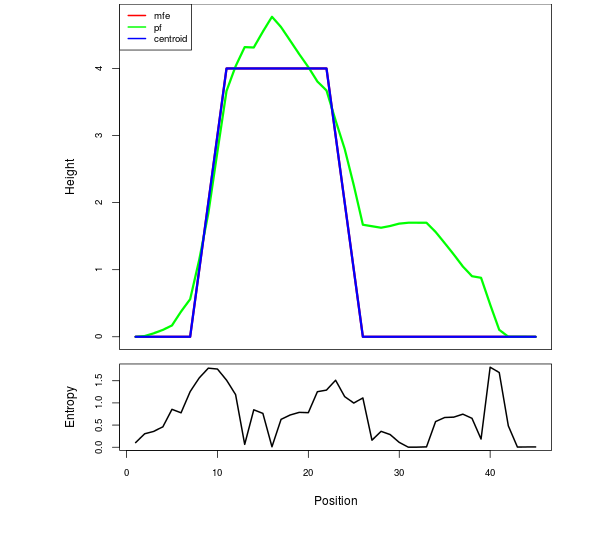
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria gonorrhoeae, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
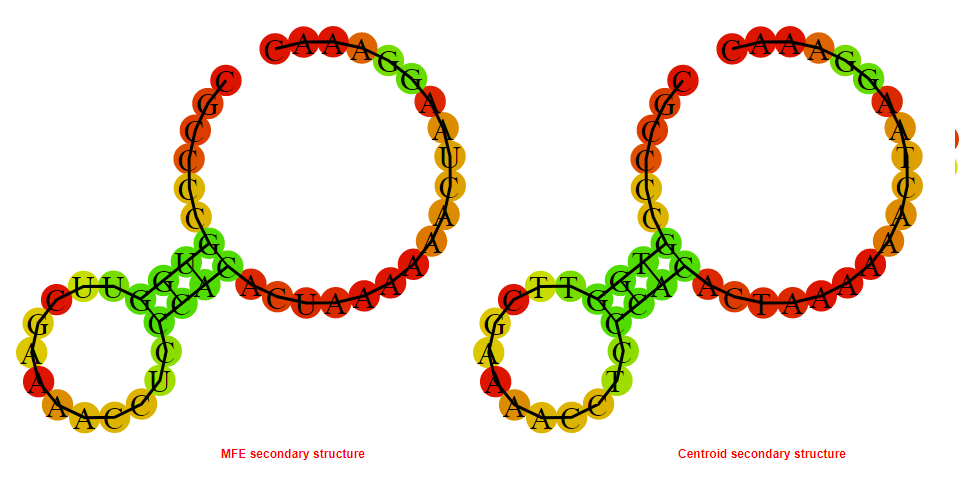
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria gonorrhoeae, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
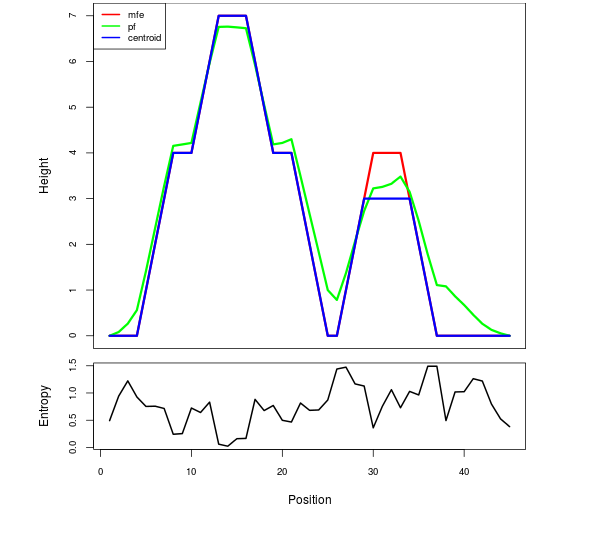
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria meningitidis, PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
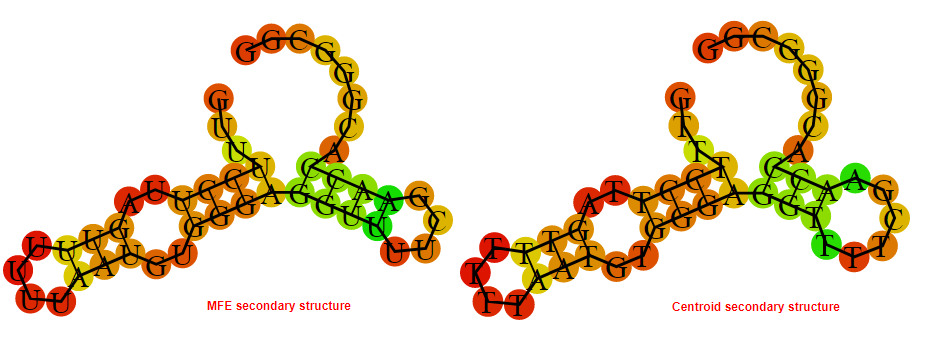
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria meningitidis, PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
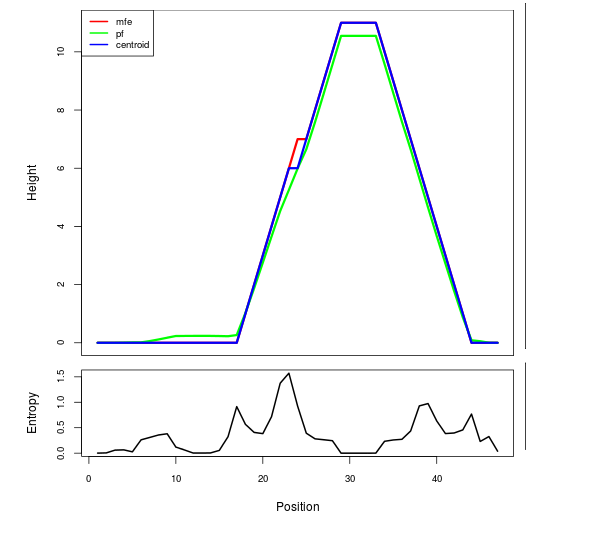
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
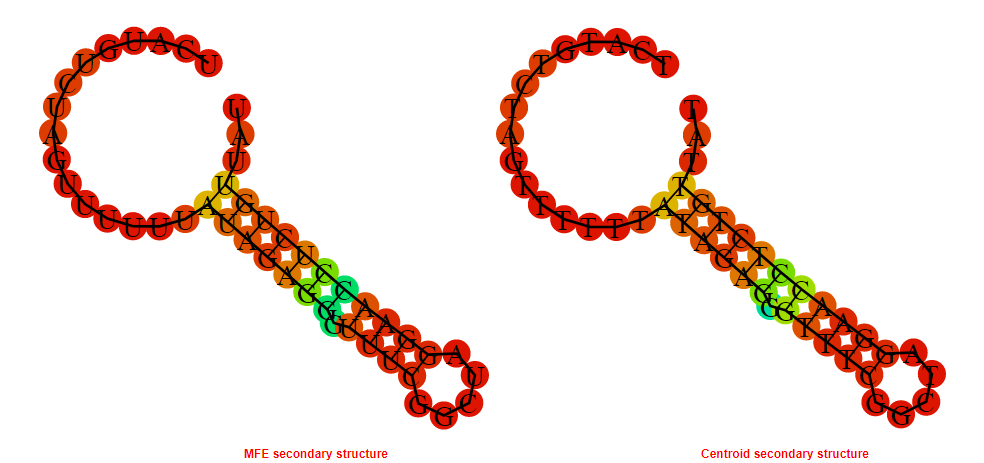
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
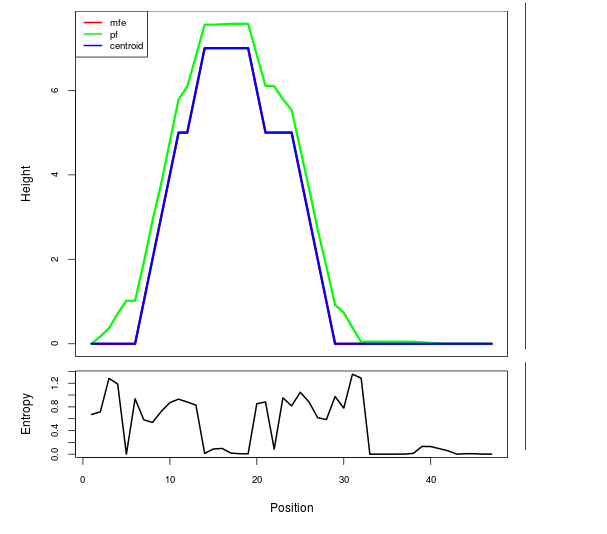
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus saprophyticus , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus saprophyticus , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
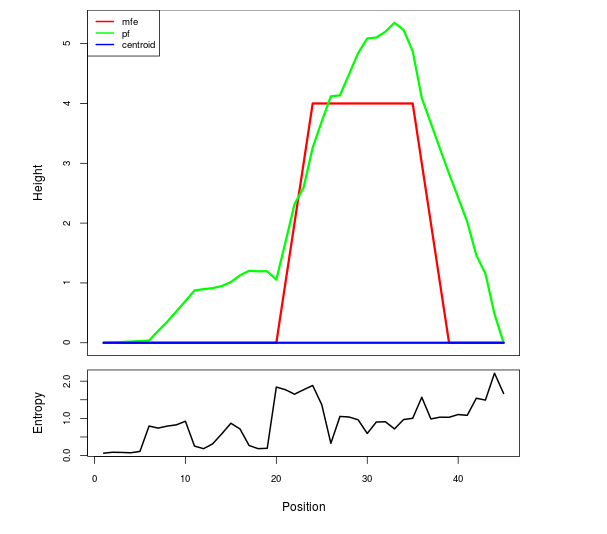
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus agalactiae , PreQ1 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
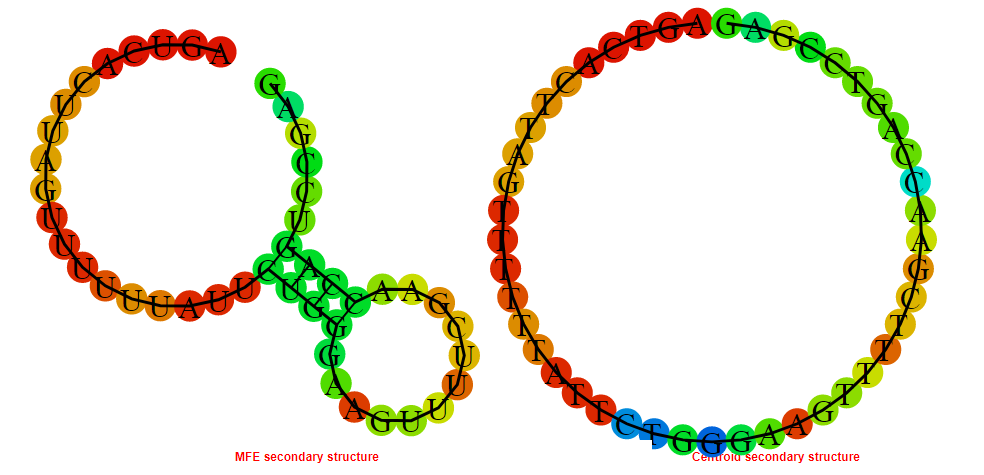
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus agalactiae , PreQ1 type switch.
Multiple alignment of the PreQ1 type switches
Below is presented the sequences alignment of all the PreQ1 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the PreQ1 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the PreQ1 type switches involved in.

Multi_resistance strains for the PreQ1 type switches
The table displays known human pathogenic bacteria bearing the PreQ1 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
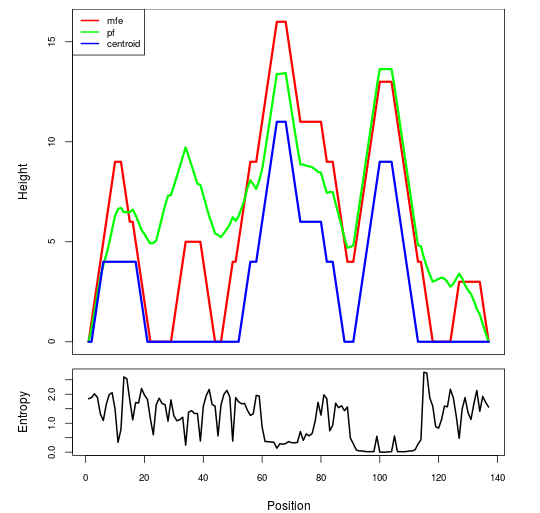
The plot represents the thermodynamic ensemble of the RNA structure of the Acinetobacter baumannii , Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
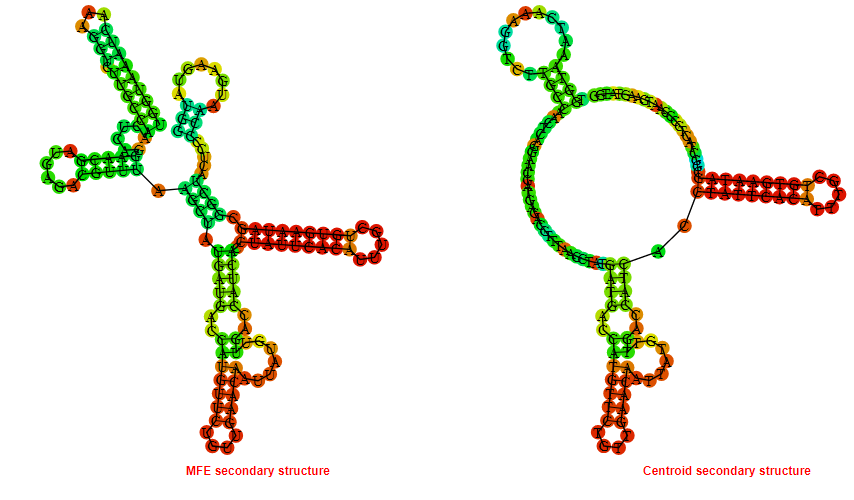
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Acinetobacter baumannii , Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.
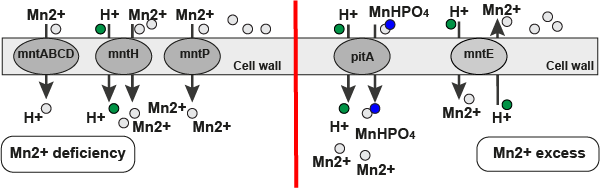
Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
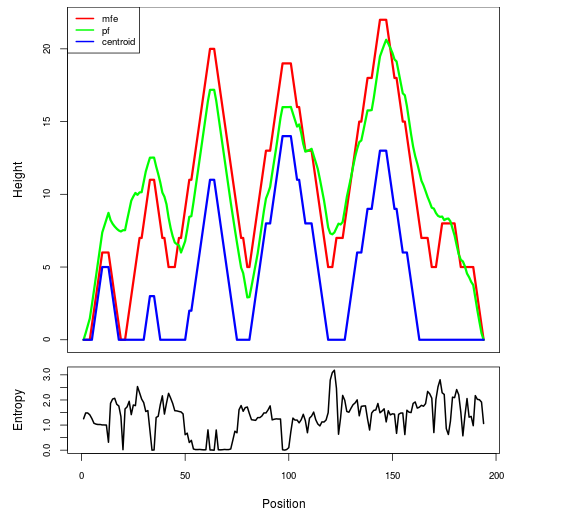
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis , Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
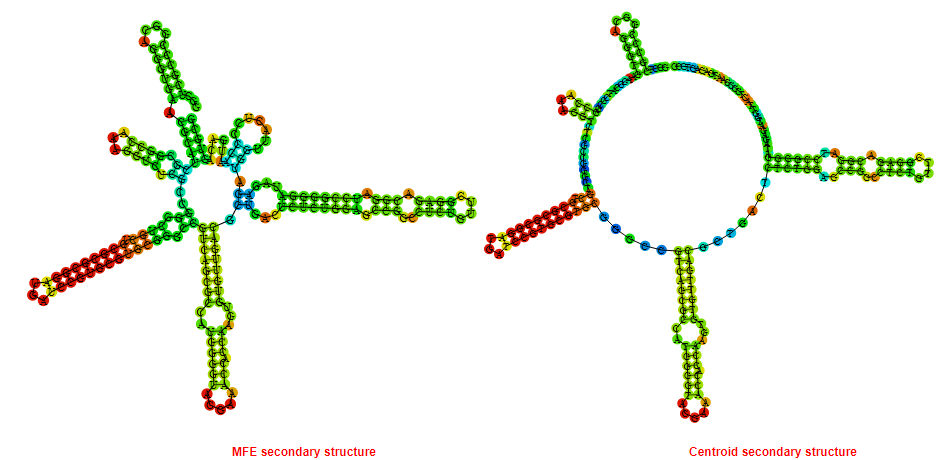
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis , Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
The plot represents the thermodynamic ensemble of the RNA structure of the Enterobacter sp, Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
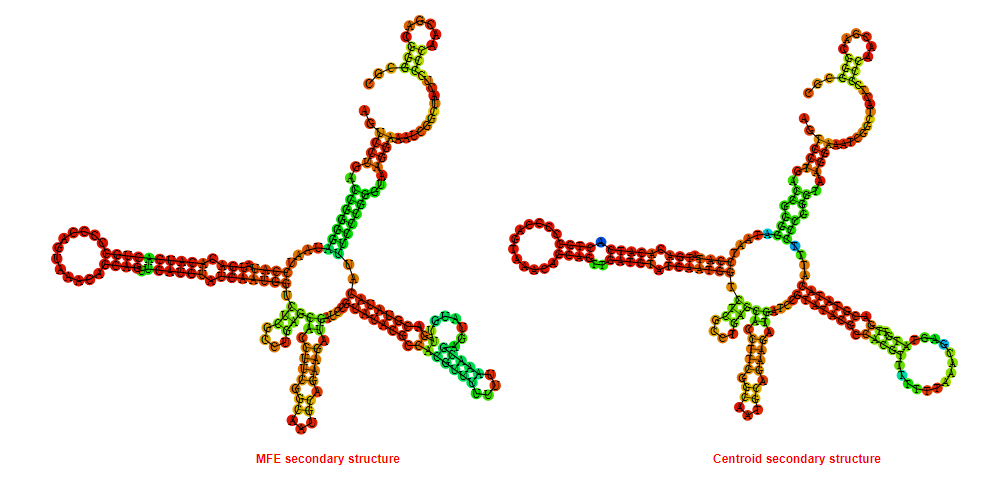
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterobacter sp, Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
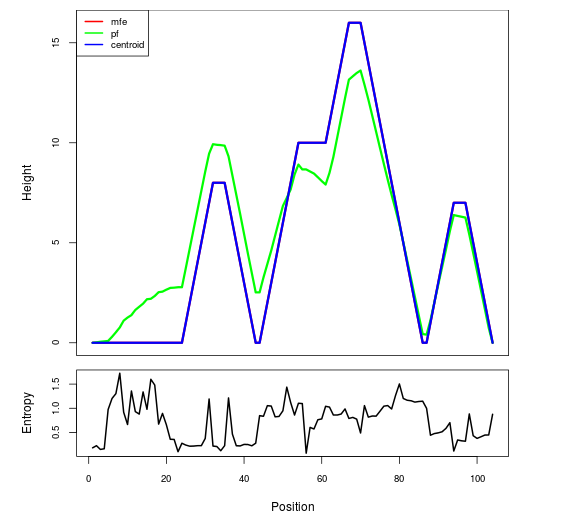
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes, Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
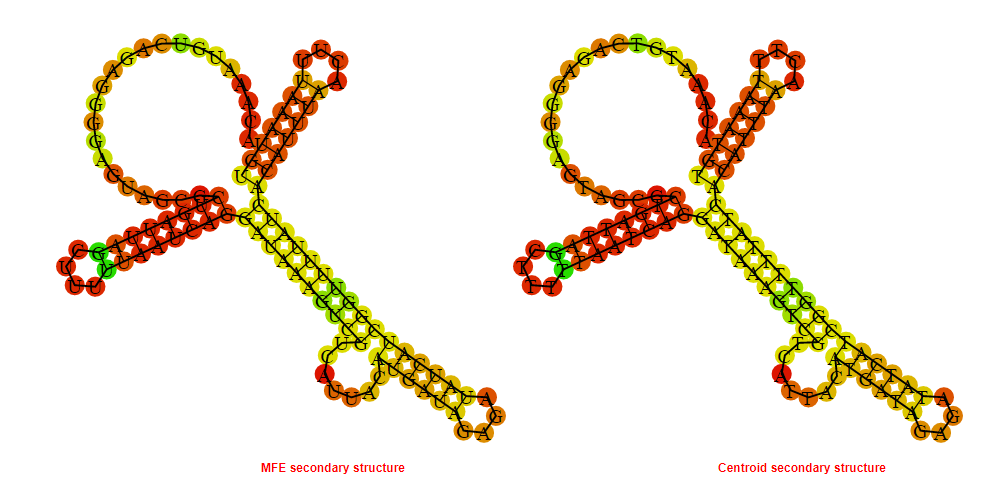
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes, Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
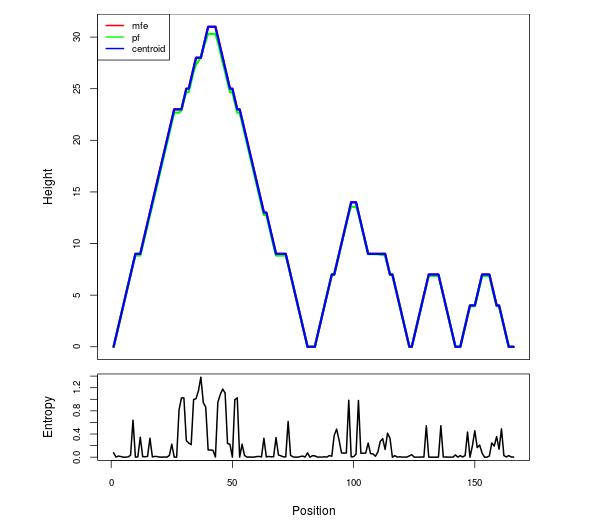
The plot represents the thermodynamic ensemble of the RNA structure of the Pseudomonas aeruginosa, Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
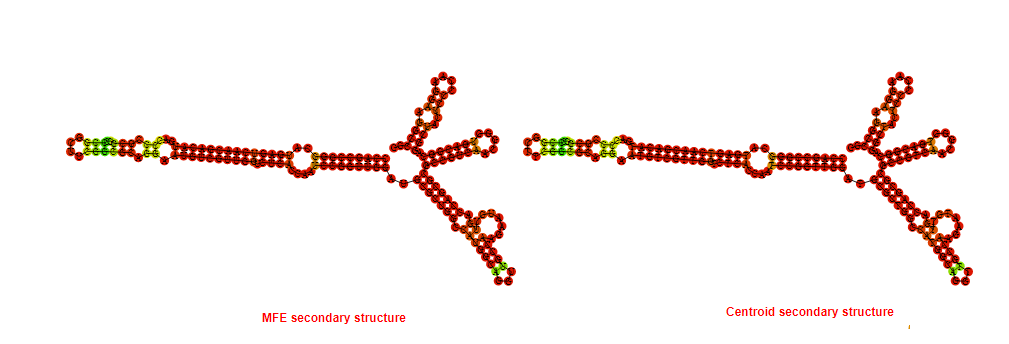
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Pseudomonas aeruginosa, Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
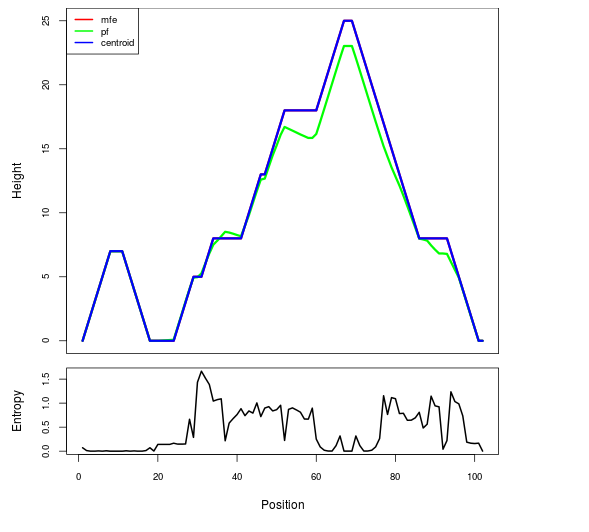
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella enterica, Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
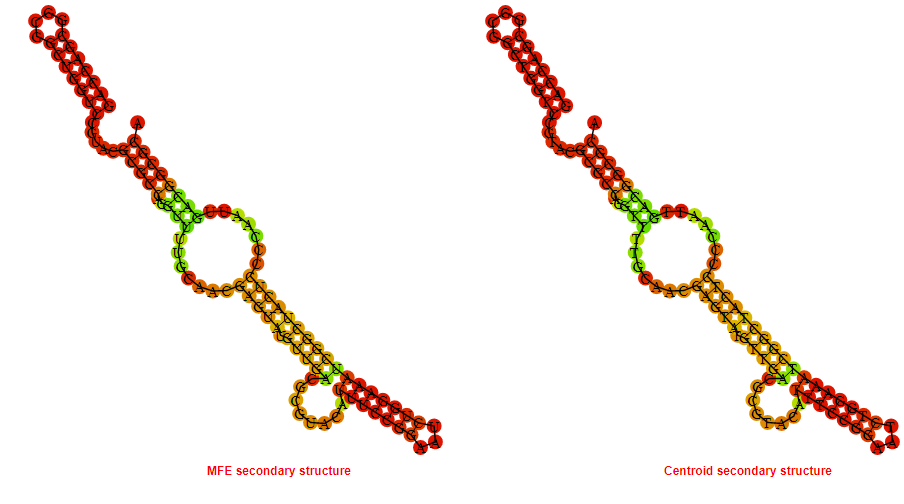
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella enterica, Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
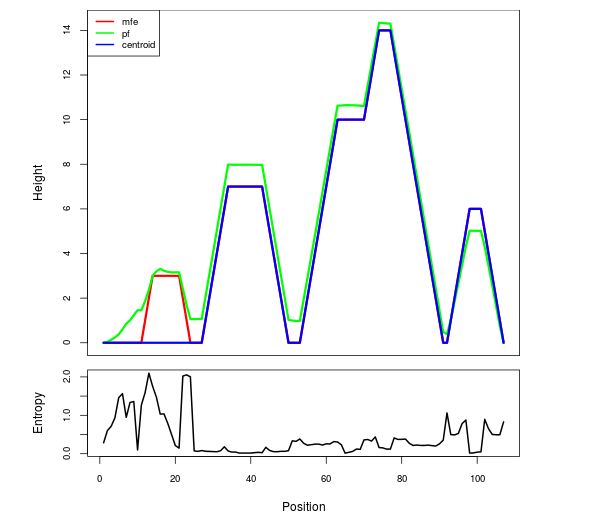
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus , Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
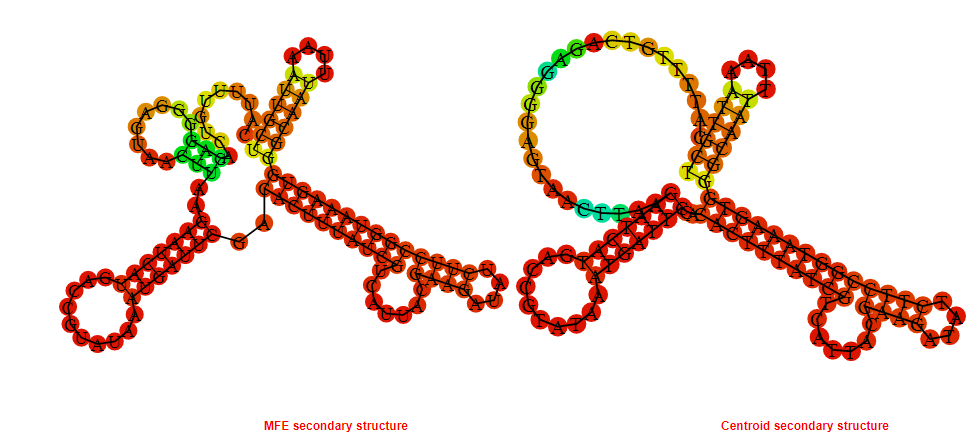
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus , Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
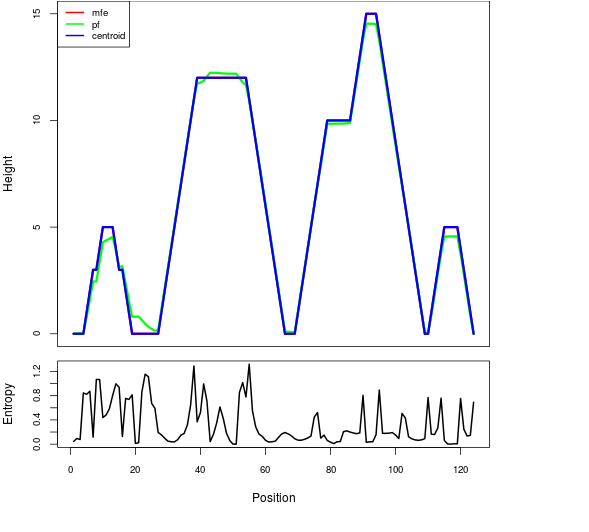
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis , Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
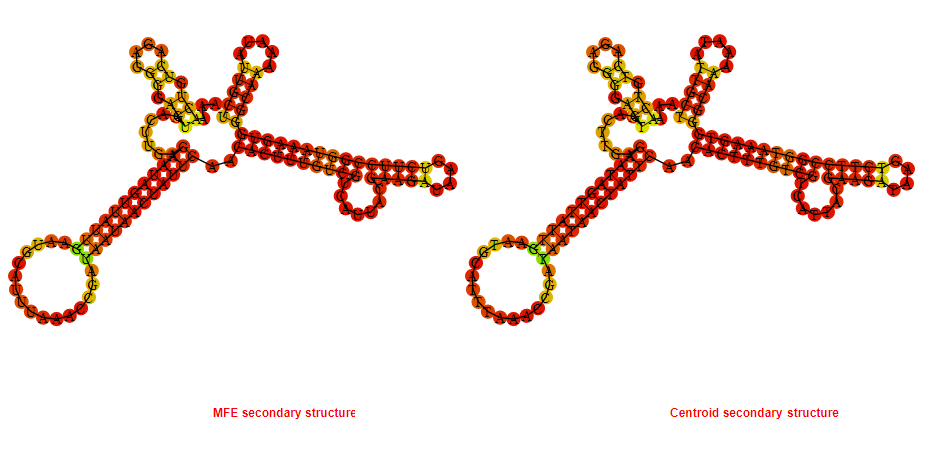
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis , Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
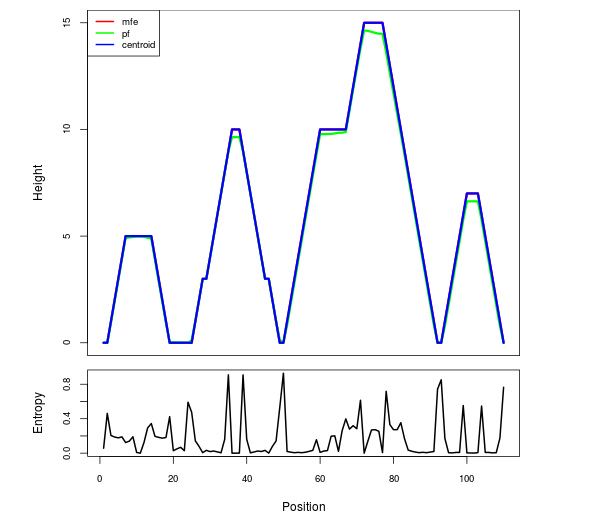
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae , Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
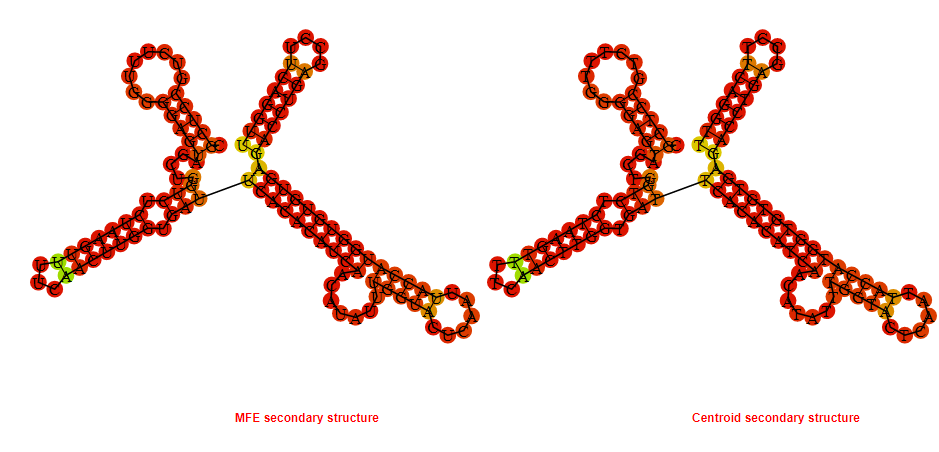
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae , Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
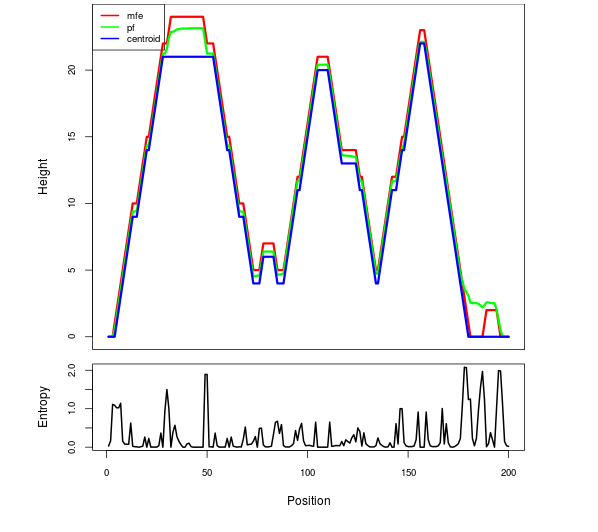
The plot represents the thermodynamic ensemble of the RNA structure of the Yersinia pestis, Mn+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
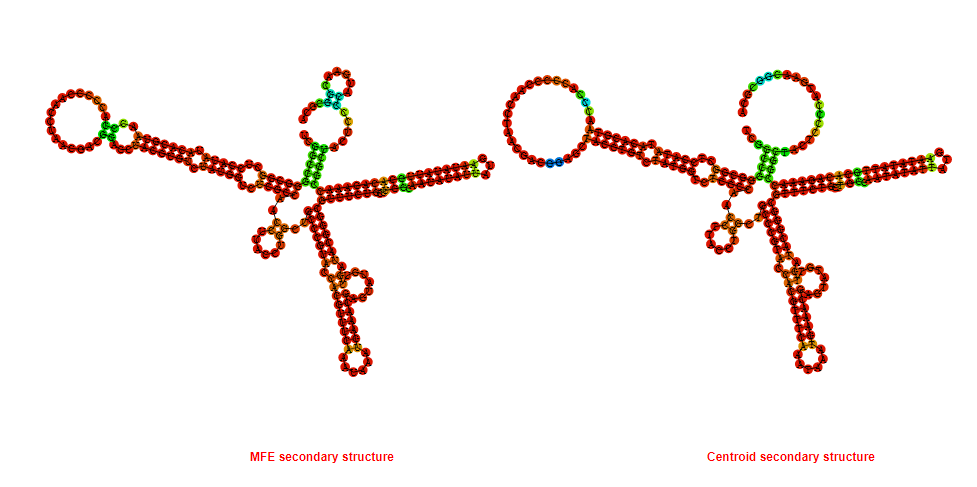
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Yersinia pestis, Mn+2 type switch.
Multiple alignment of the Mn+2 type switches
Below is presented the sequences alignment of all the Mn+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mn+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mn+2 type switches involved in.

Multi_resistance strains for the Mn+2 type switches
The table displays known human pathogenic bacteria bearing the Mn+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
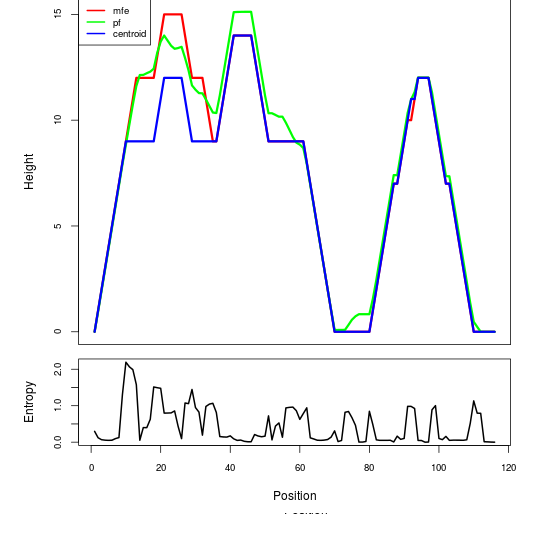
The plot represents the thermodynamic ensemble of the RNA structure of the Escherichia coli, Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
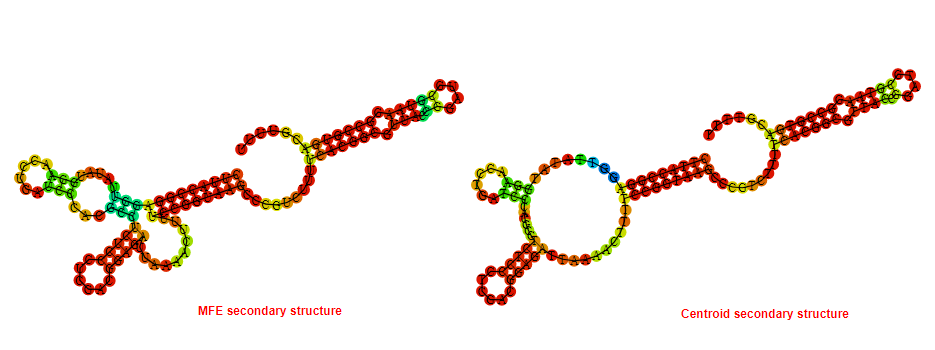
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Escherichia coli, Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.
Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
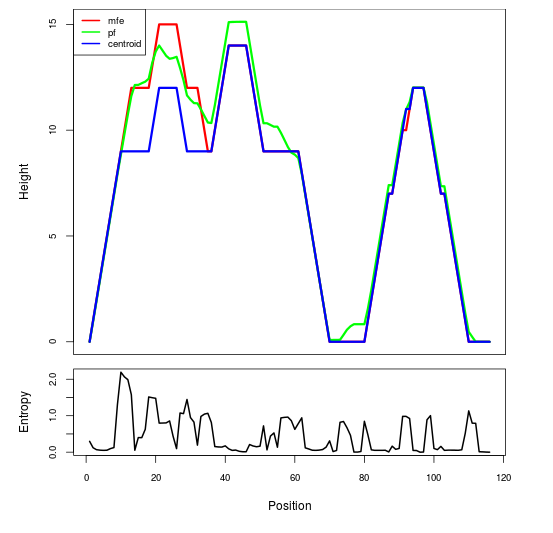
The plot represents the thermodynamic ensemble of the RNA structure of the Shigella dysenteriae , Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
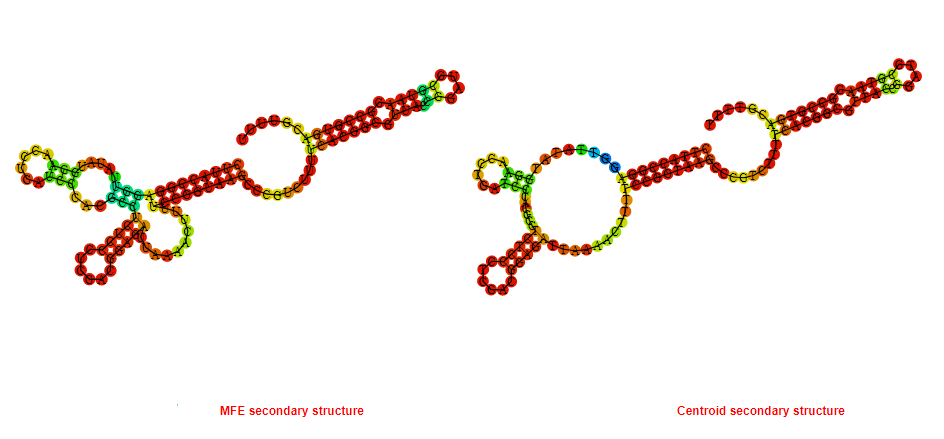
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Shigella dysenteriae , Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.

Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
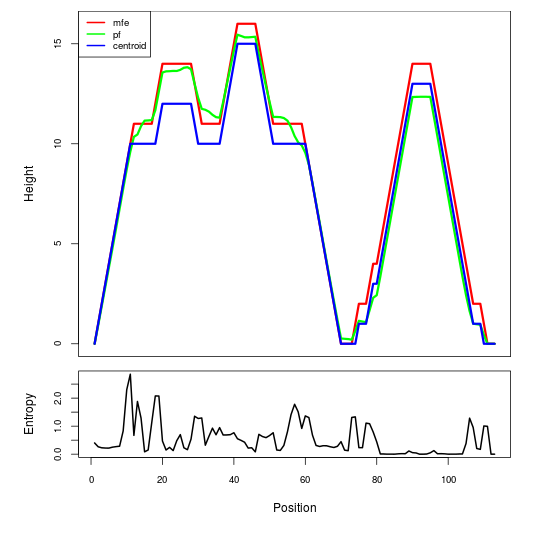
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella Typhimurium , Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
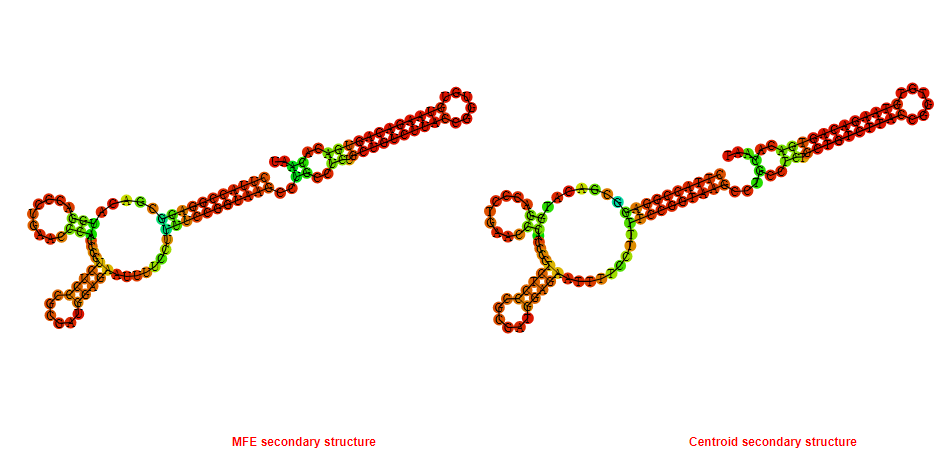
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella Typhimurium , Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.

Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
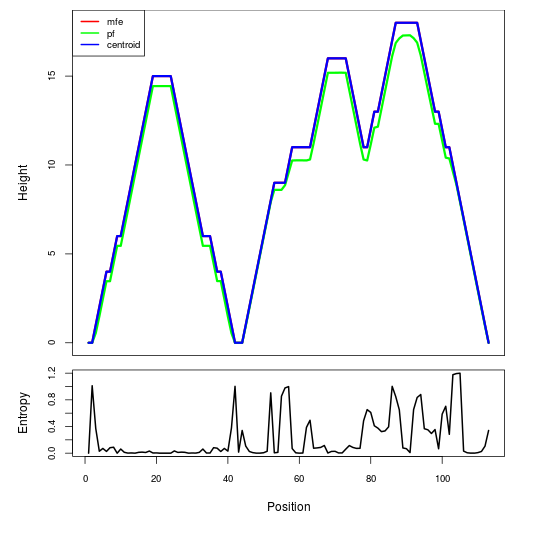
The plot represents the thermodynamic ensemble of the RNA structure of the Salmonella enterica, Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
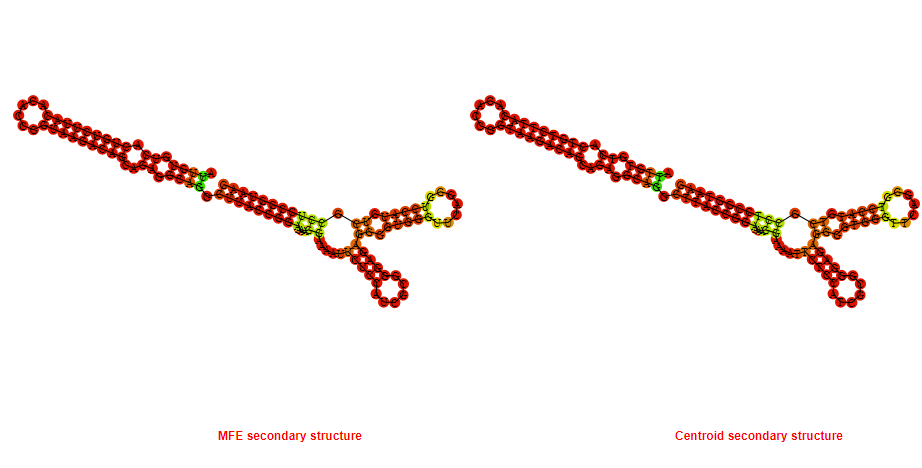
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Salmonella enterica, Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.

Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
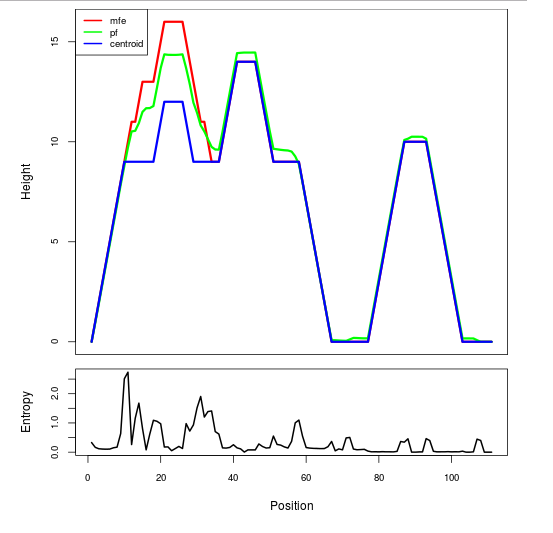
The plot represents the thermodynamic ensemble of the RNA structure of the Enterobacter , Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
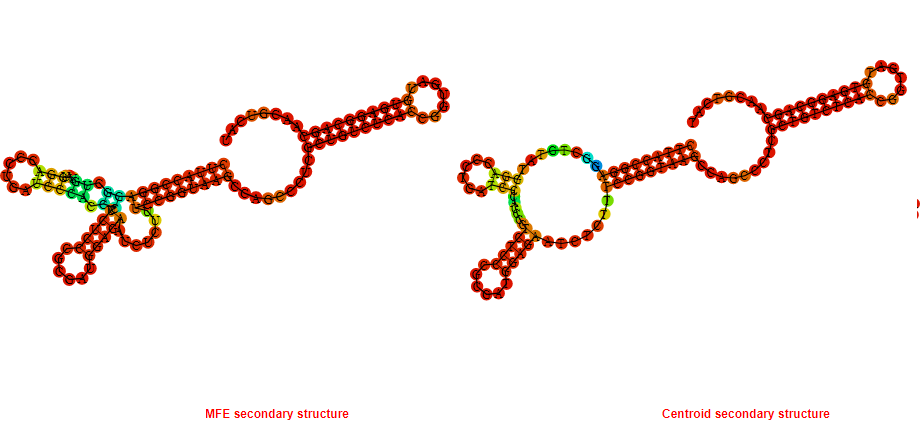
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterobacter , Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.

Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
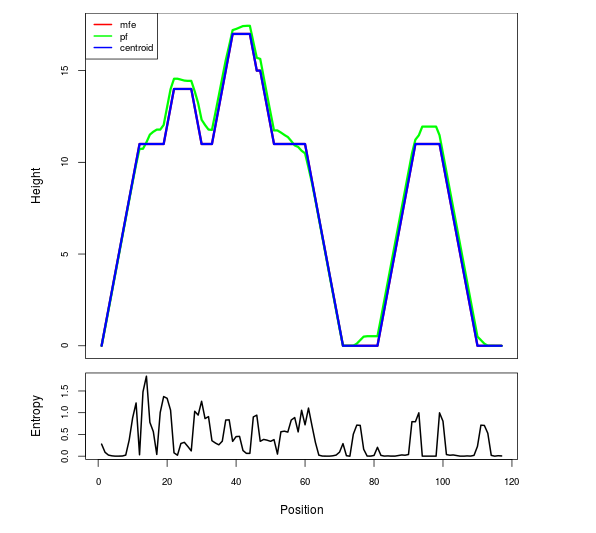
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae, Mg+2 type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
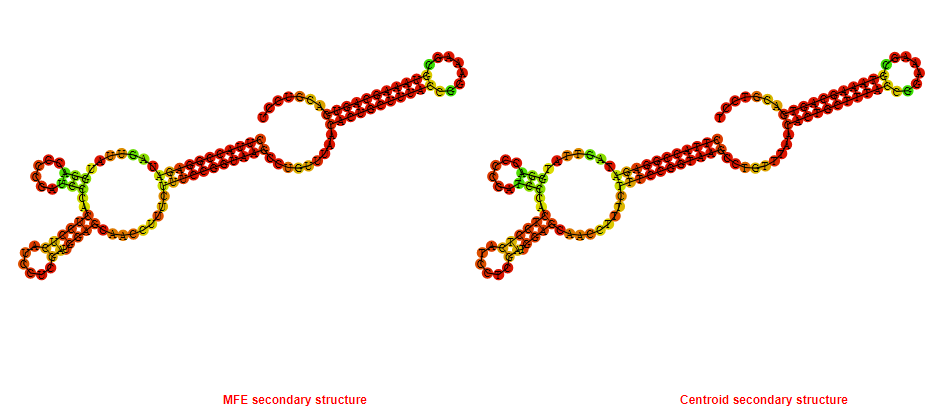
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae, Mg+2 type switch.
Multiple alignment of the Mg+2 type switches
Below is presented the sequences alignment of all the Mg+2 type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Mg+2 type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Mg+2 type switches involved in.

Multi_resistance strains for the Mg+2 type switches
The table displays known human pathogenic bacteria bearing the Mg+2 type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
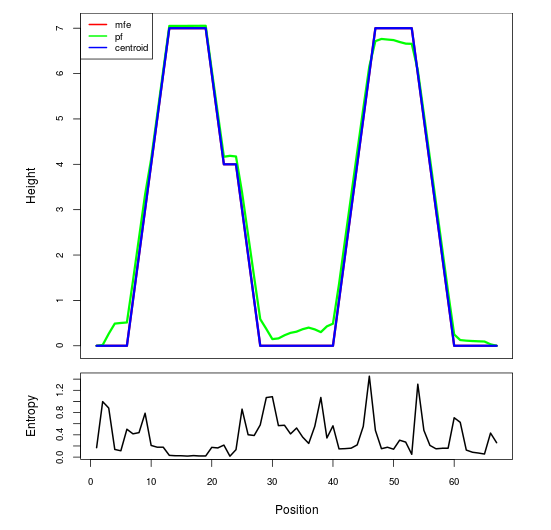
The plot represents the thermodynamic ensemble of the RNA structure of the Acinetobacter baumannii , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
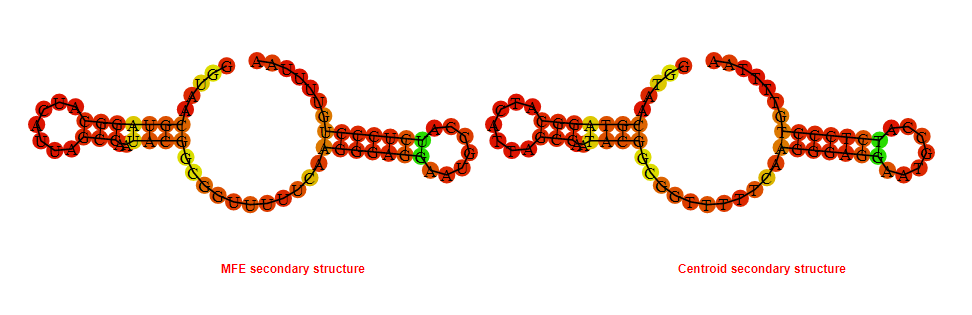
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Acinetobacter baumannii , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.
Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
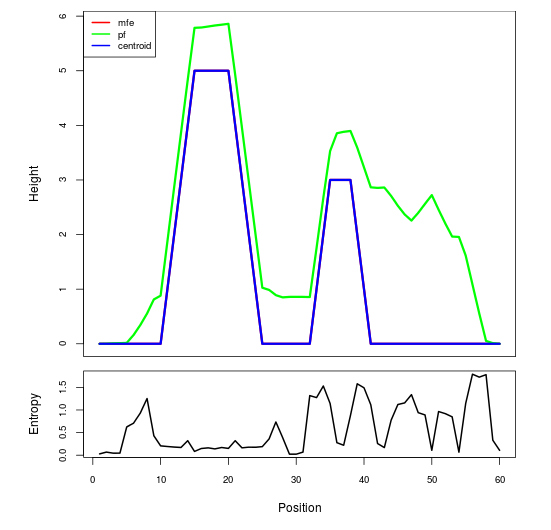
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
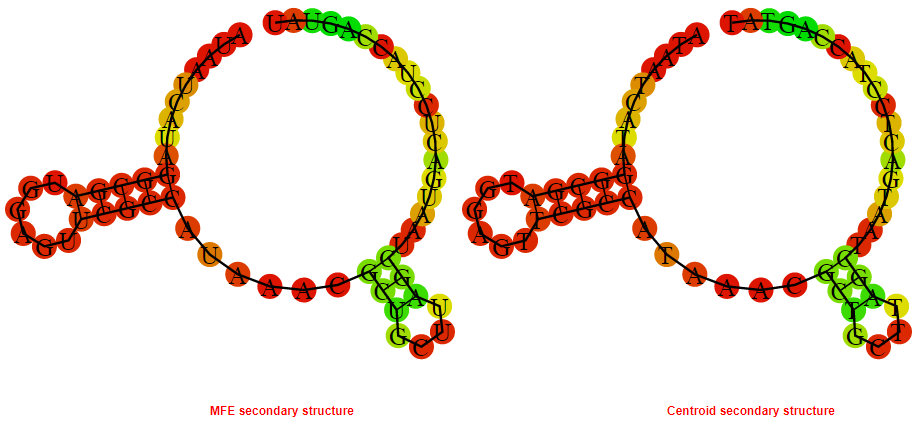
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
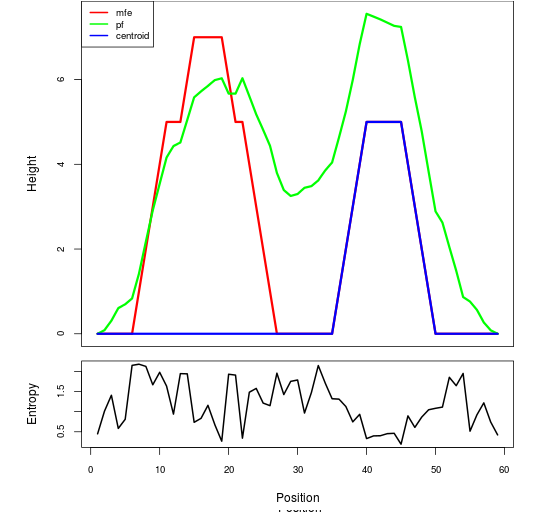
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
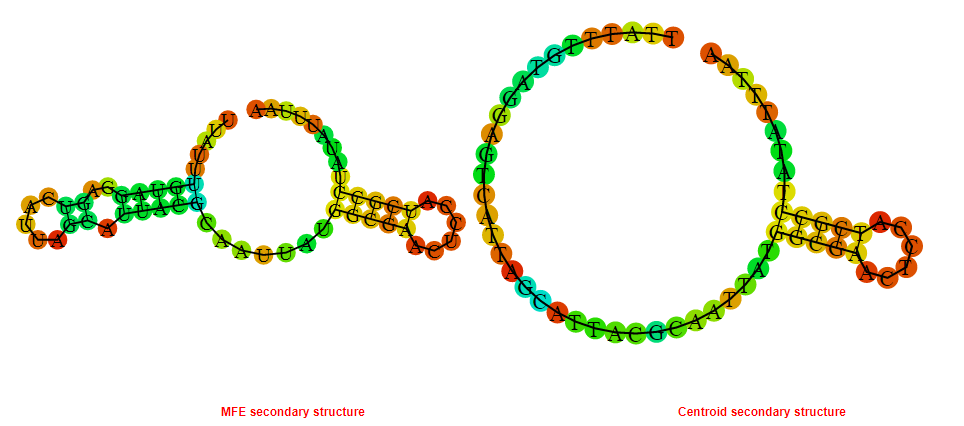
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
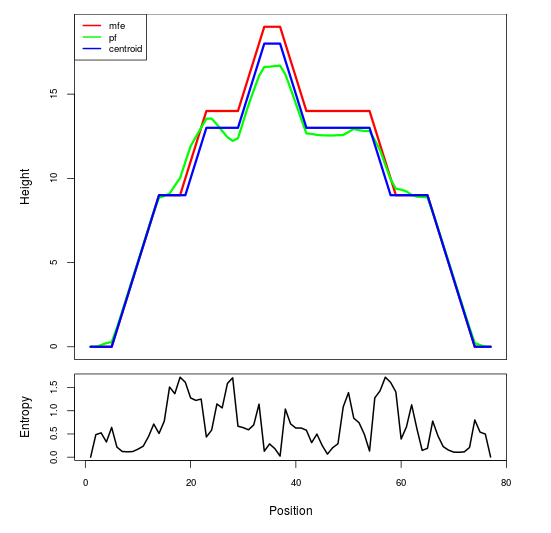
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium difficile , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
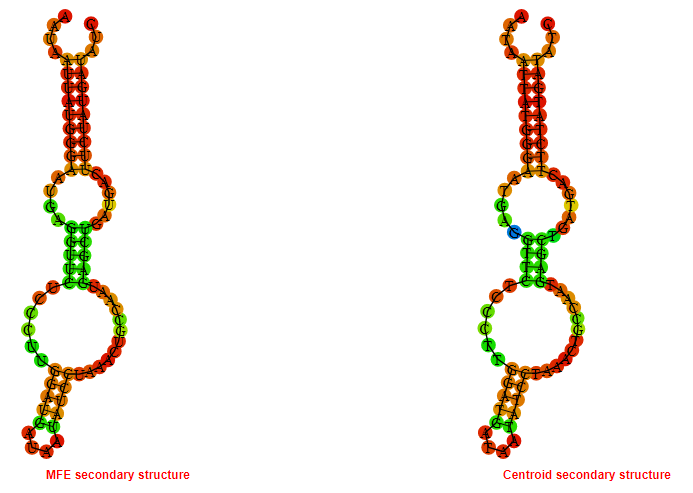
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium difficile , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
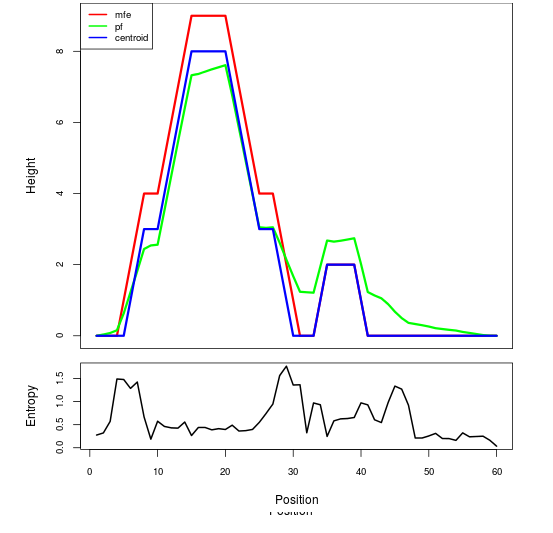
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
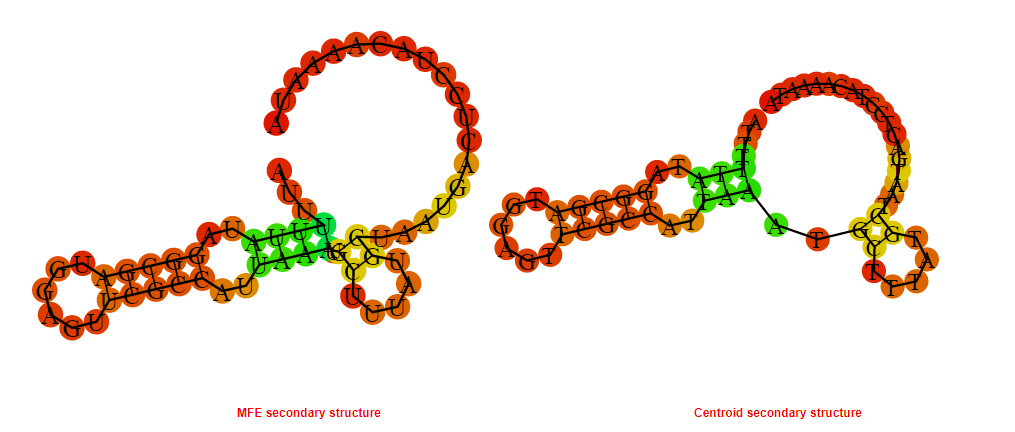
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
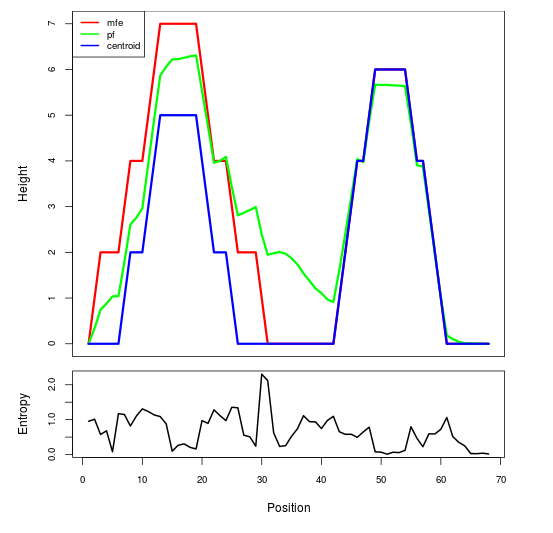
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecalis , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
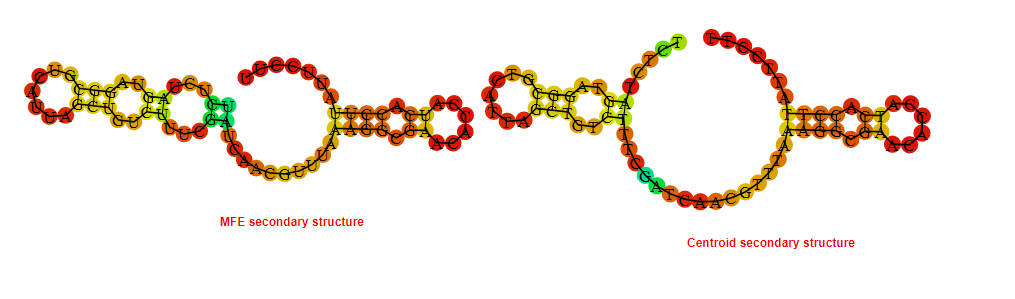
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecalis , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
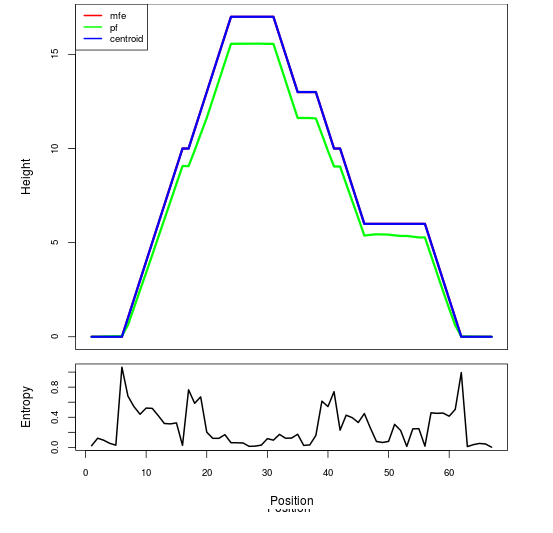
The plot represents the thermodynamic ensemble of the RNA structure of the Enterococcus faecium , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
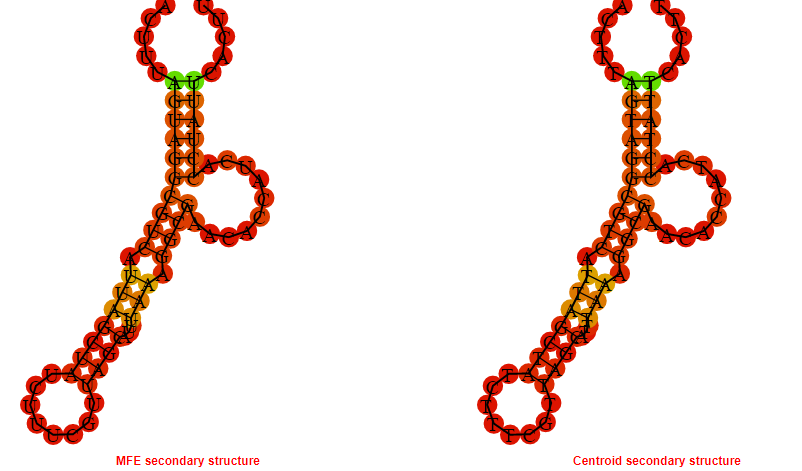
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Enterococcus faecium , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
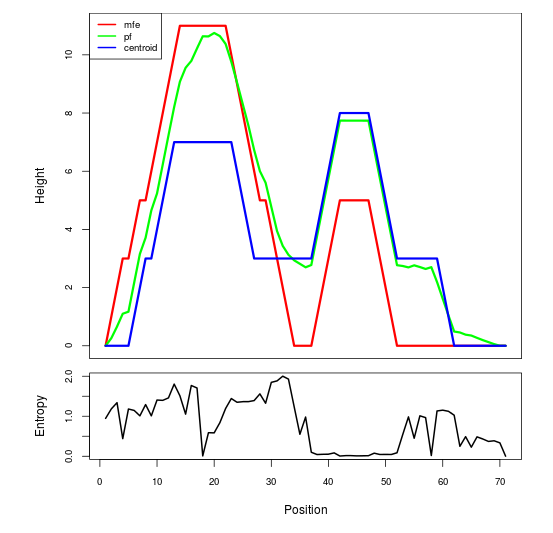
The plot represents the thermodynamic ensemble of the RNA structure of the Klebsiella pneumoniae , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
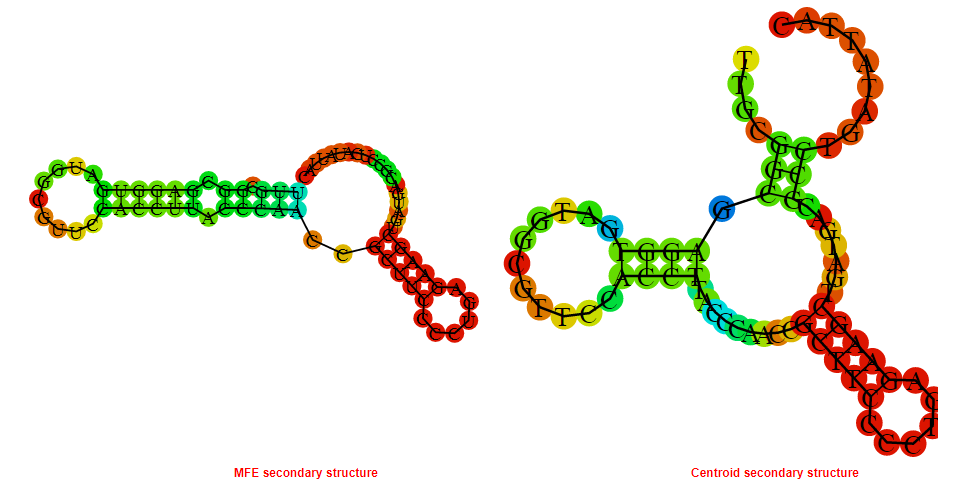
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Klebsiella pneumoniae , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
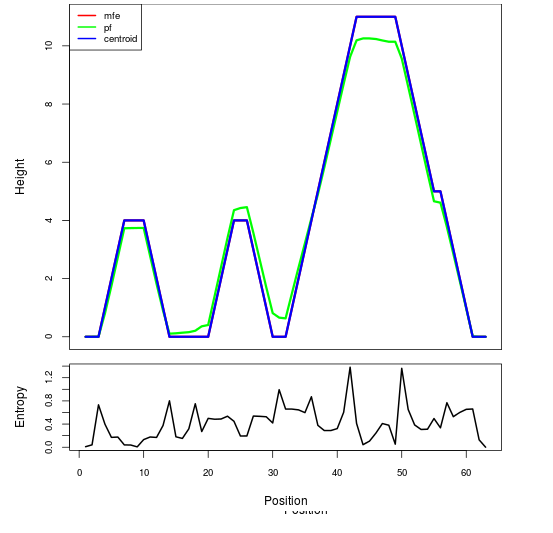
The plot represents the thermodynamic ensemble of the RNA structure of the Pseudomonas aeruginosa , Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
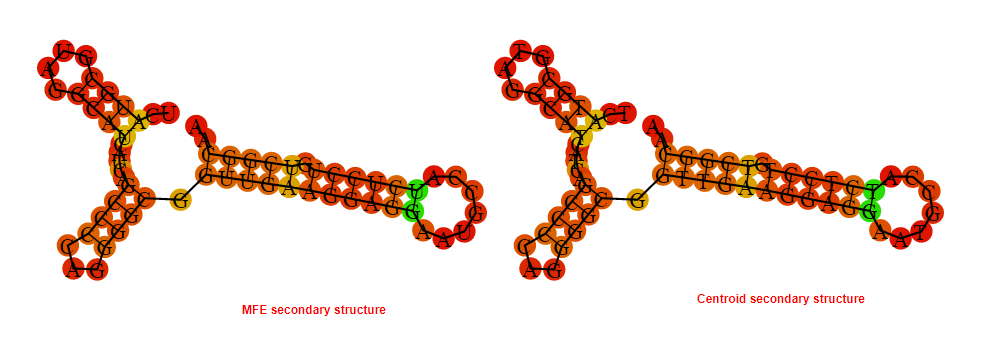
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Pseudomonas aeruginosa , Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
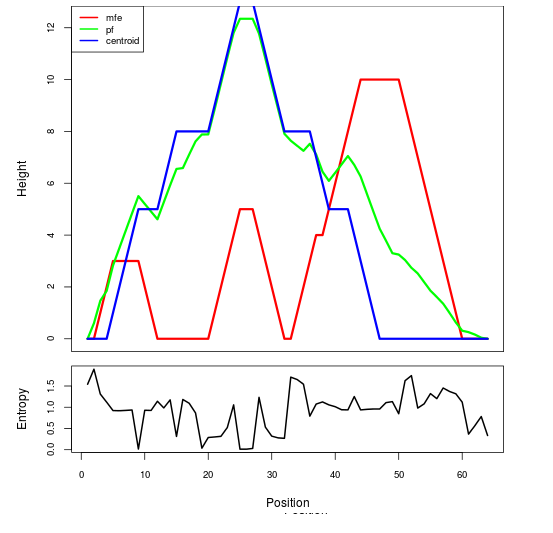
The plot represents the thermodynamic ensemble of the RNA structure of the Yersinia pestis, Fluoride type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
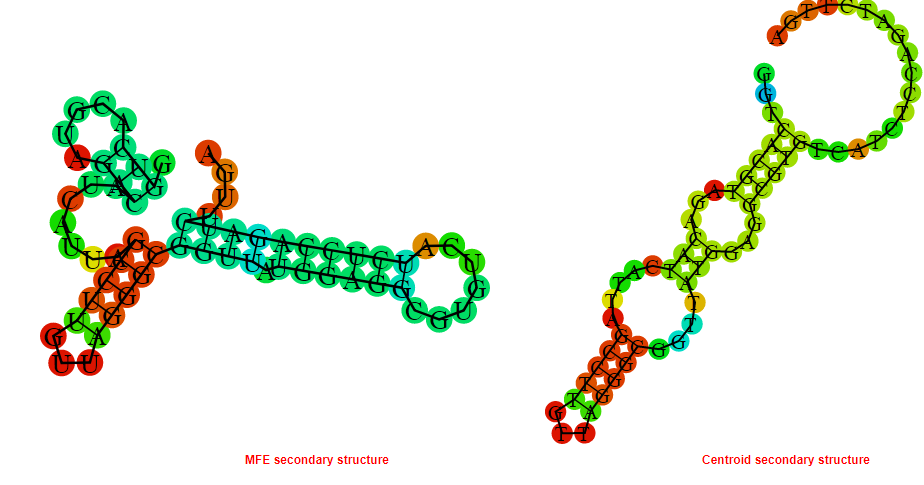
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Yersinia pestis, Fluoride type switch.
Multiple alignment of the Fluoride type switches
Below is presented the sequences alignment of all the Fluoride type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Fluoride type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Fluoride type switches involved in.

Multi_resistance strains for the Fluoride type switches
The table displays known human pathogenic bacteria bearing the Fluoride type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
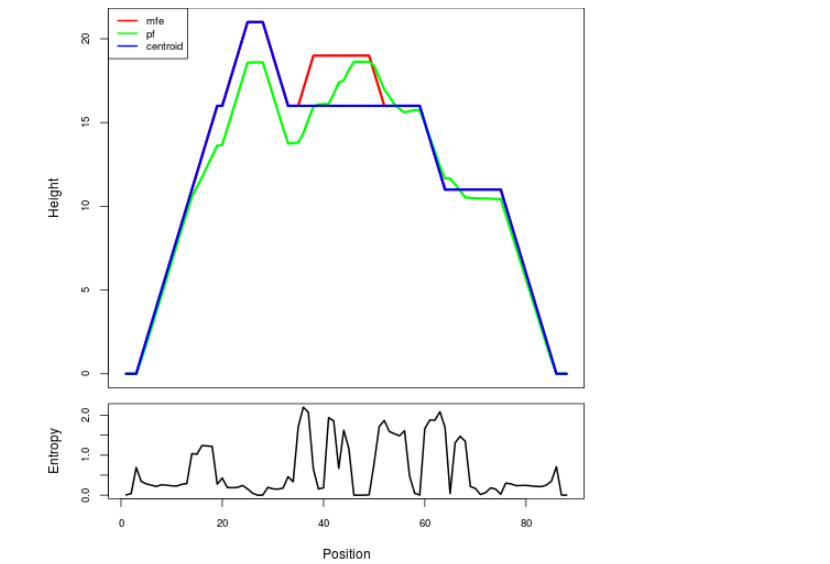
The plot represents the thermodynamic ensemble of the RNA structure of the Acinetobacter baumannii, Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
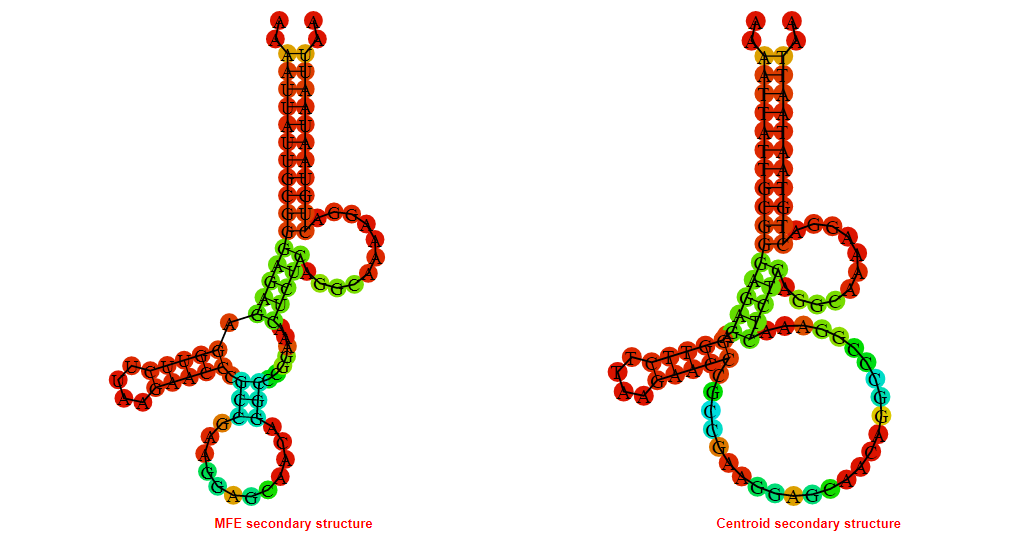
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Acinetobacter baumannii, Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.
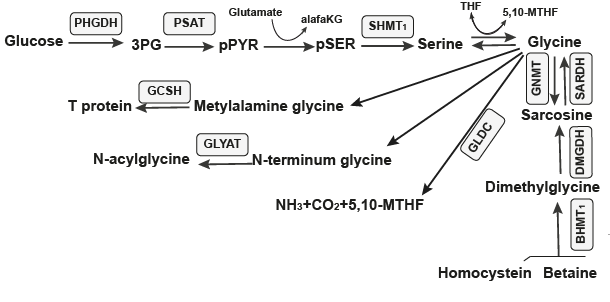
Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
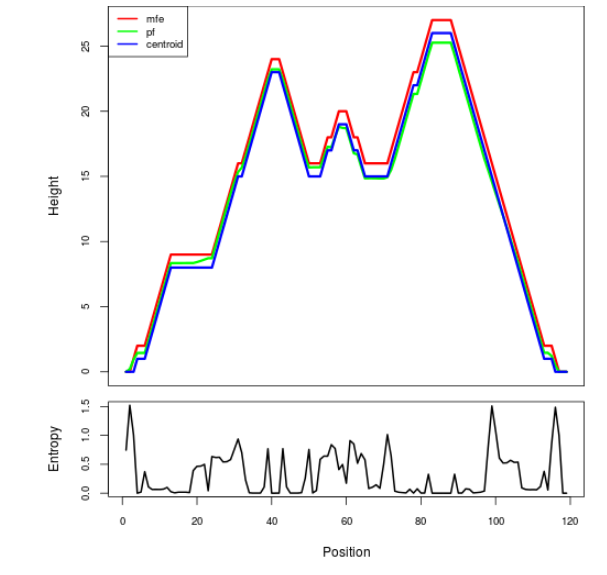
The plot represents the thermodynamic ensemble of the RNA structure of the Bacillus anthracis , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
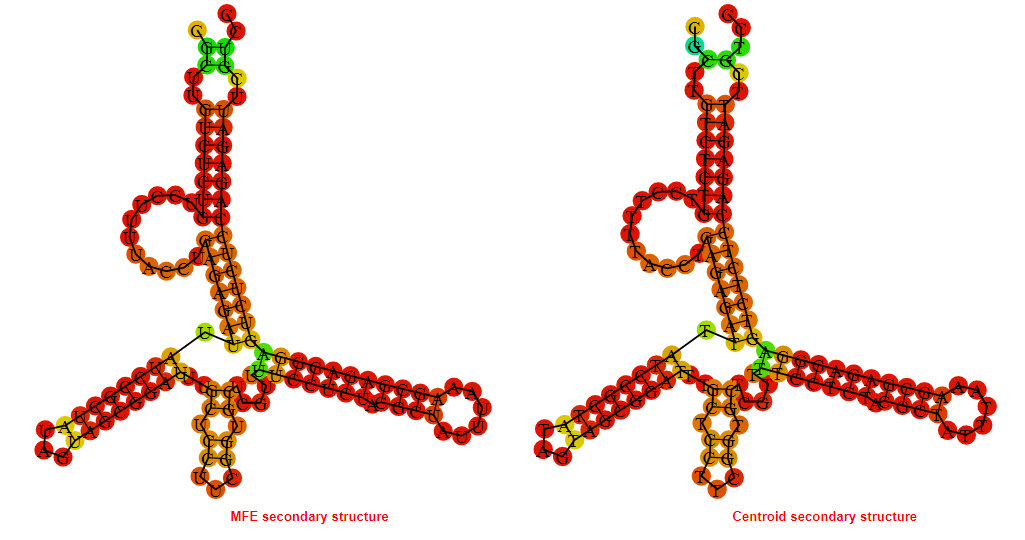
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bacillus anthracis , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
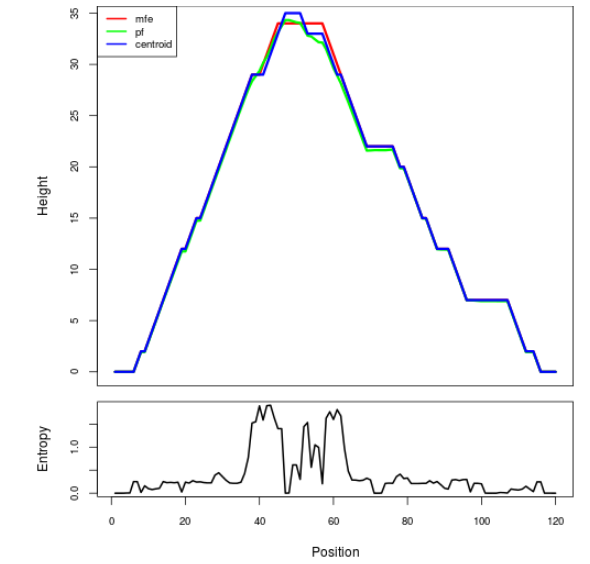
The plot represents the thermodynamic ensemble of the RNA structure of the Bordetella pertussis , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
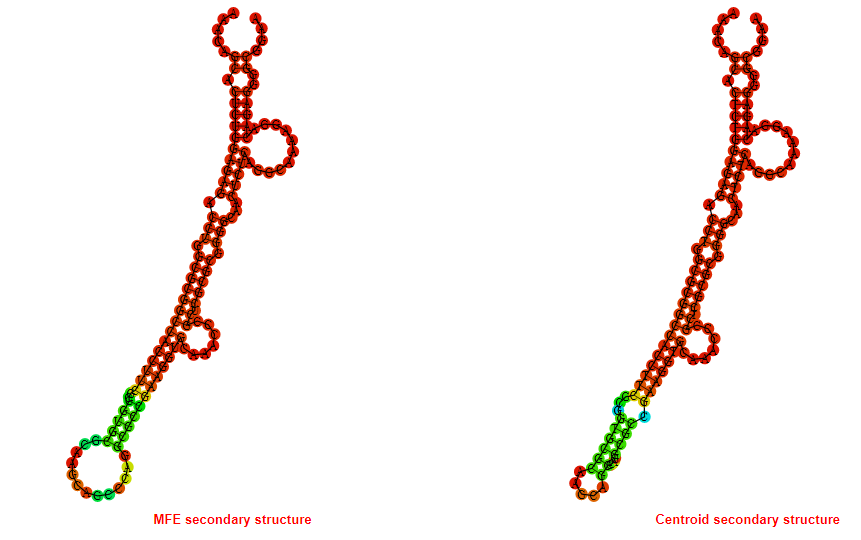
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Bordetella pertussis , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
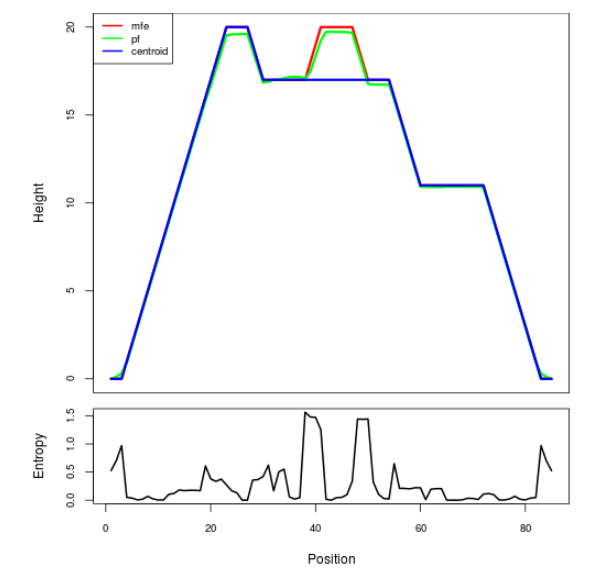
The plot represents the thermodynamic ensemble of the RNA structure of the Brucella melitensis Abortus , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
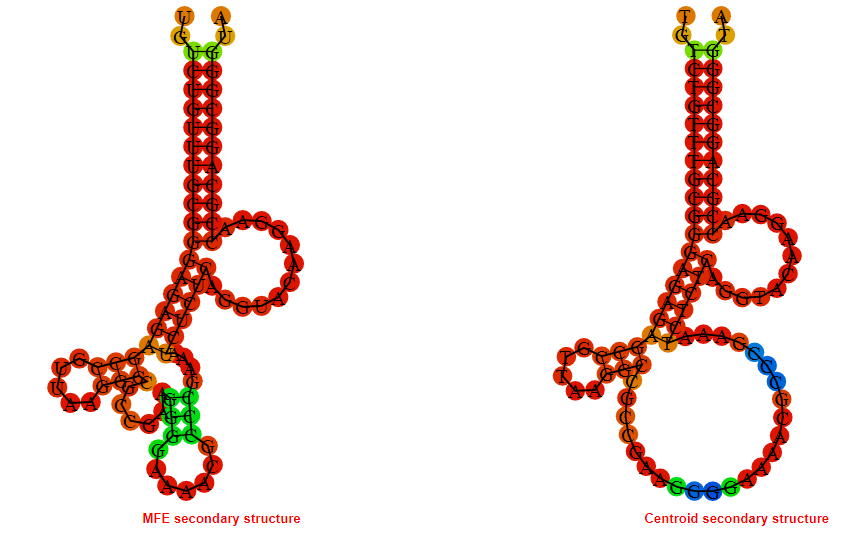
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Brucella melitensis Abortus , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
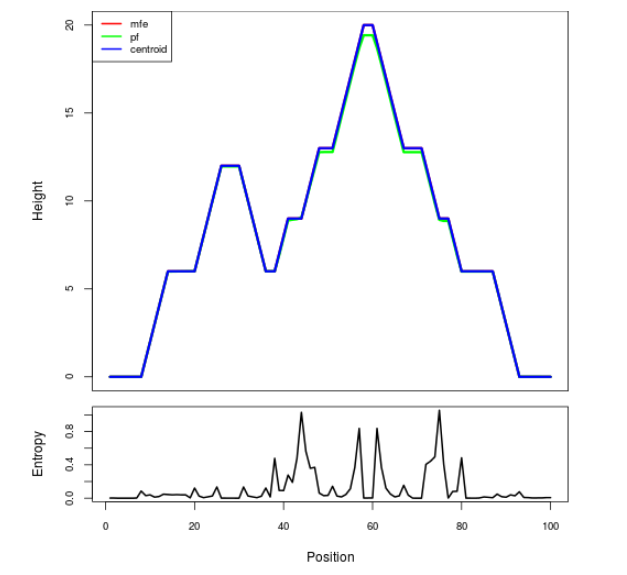
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium botulinum , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
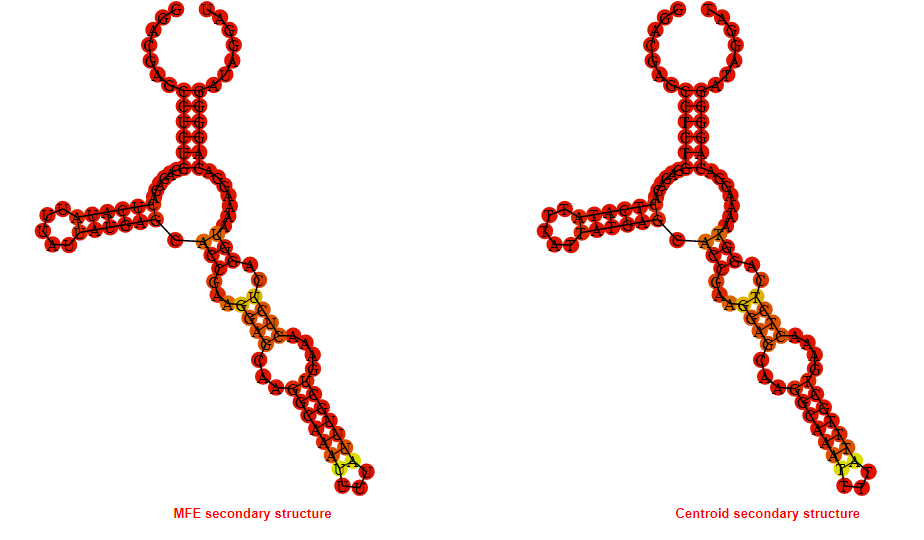
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium botulinum , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
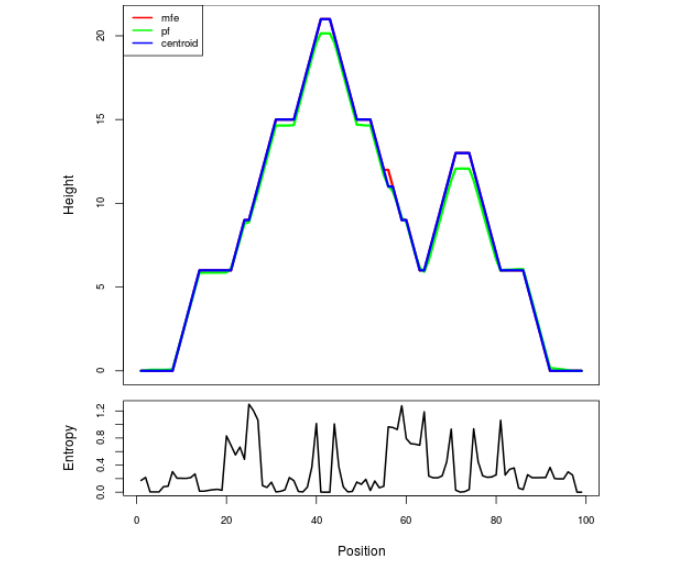
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium perfringens , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
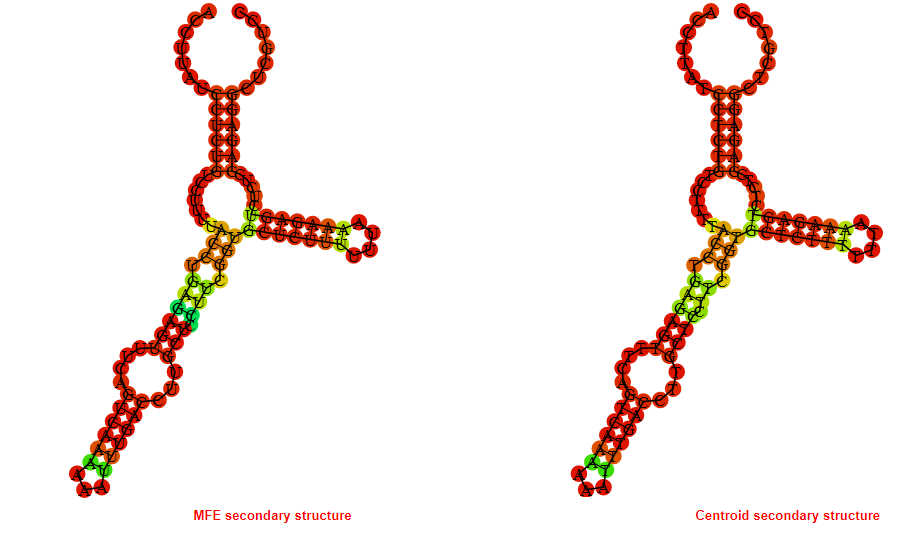
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium perfringens , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
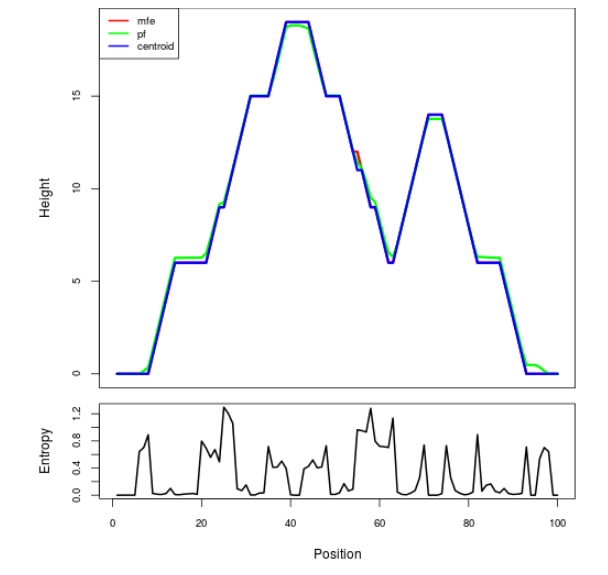
The plot represents the thermodynamic ensemble of the RNA structure of the Clostridium tetani E88 , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
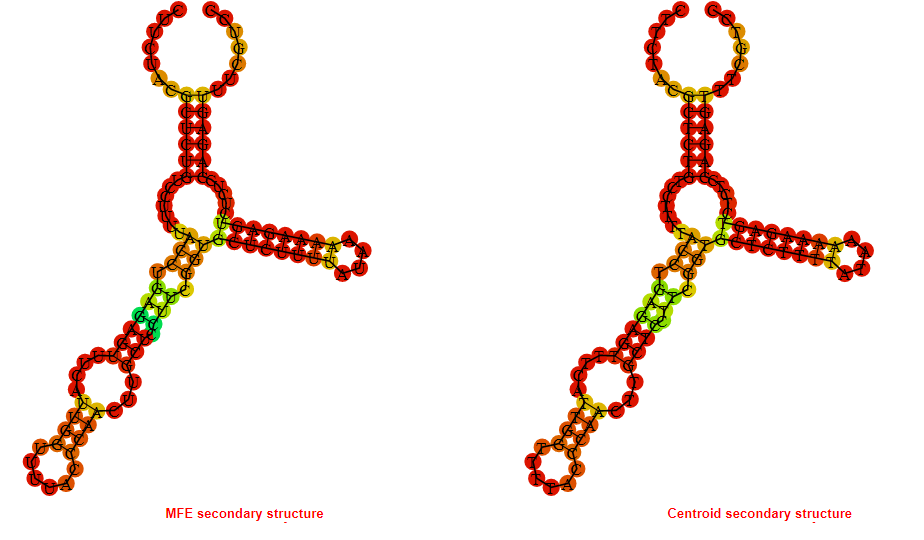
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Clostridium tetani E88 , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
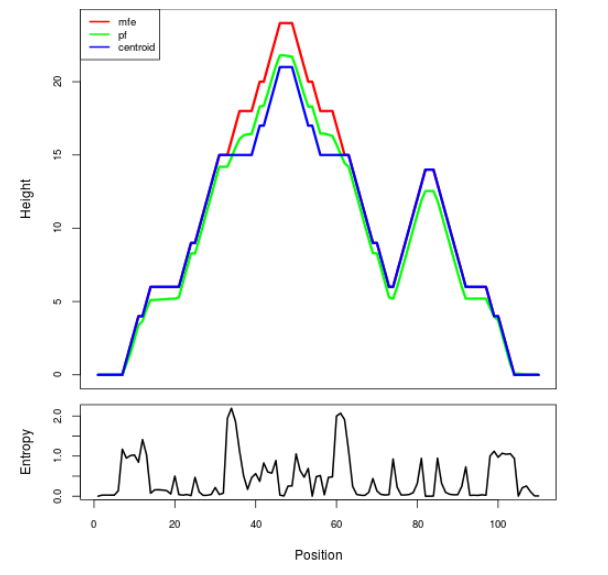
The plot represents the thermodynamic ensemble of the RNA structure of the Haemophilus influenzae , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)

Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Haemophilus influenzae , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
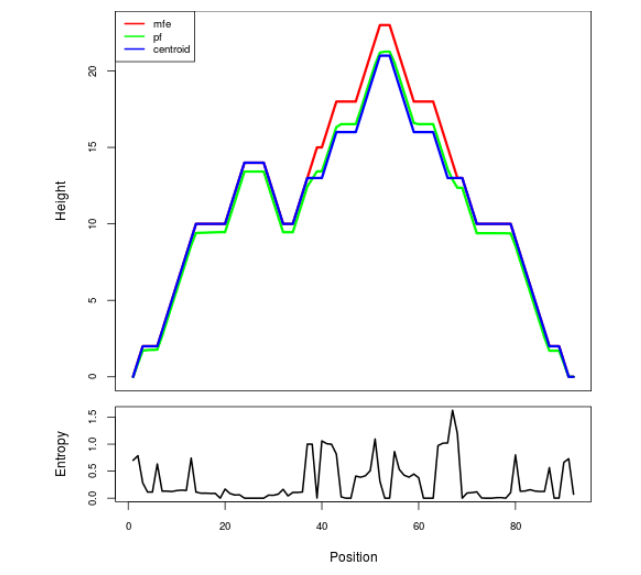
The plot represents the thermodynamic ensemble of the RNA structure of the Listeria monocytogenes , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
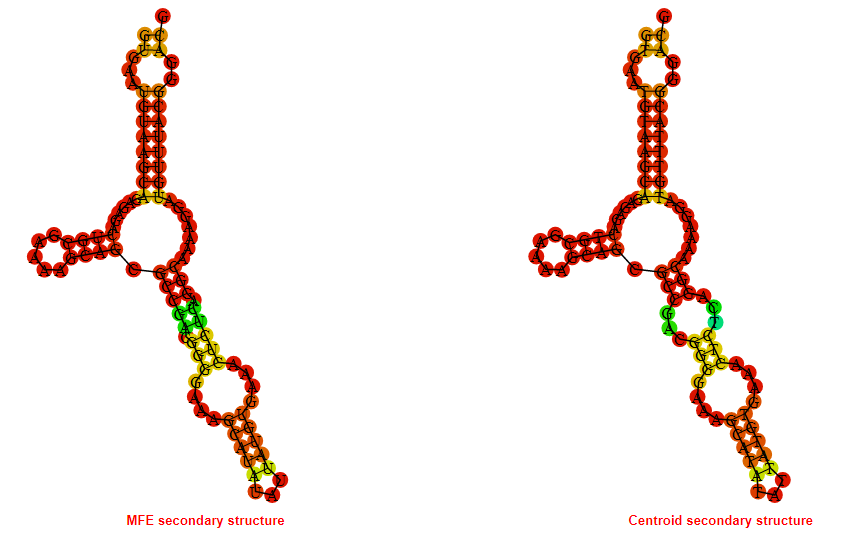
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Listeria monocytogenes , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
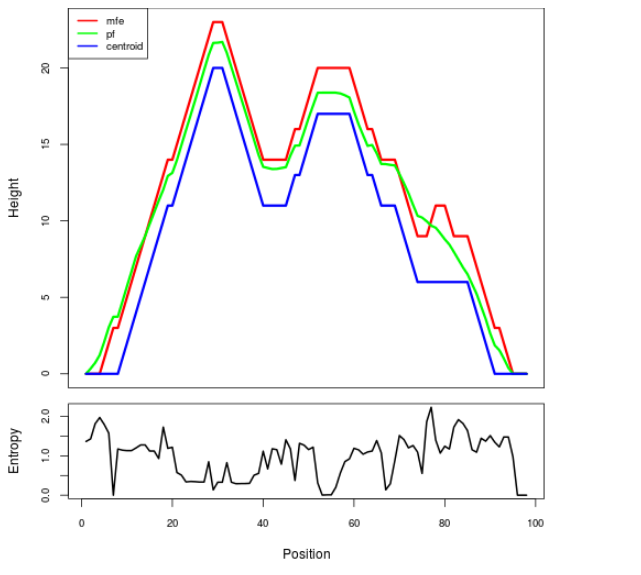
The plot represents the thermodynamic ensemble of the RNA structure of the Mycobacterium tuberculosis , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
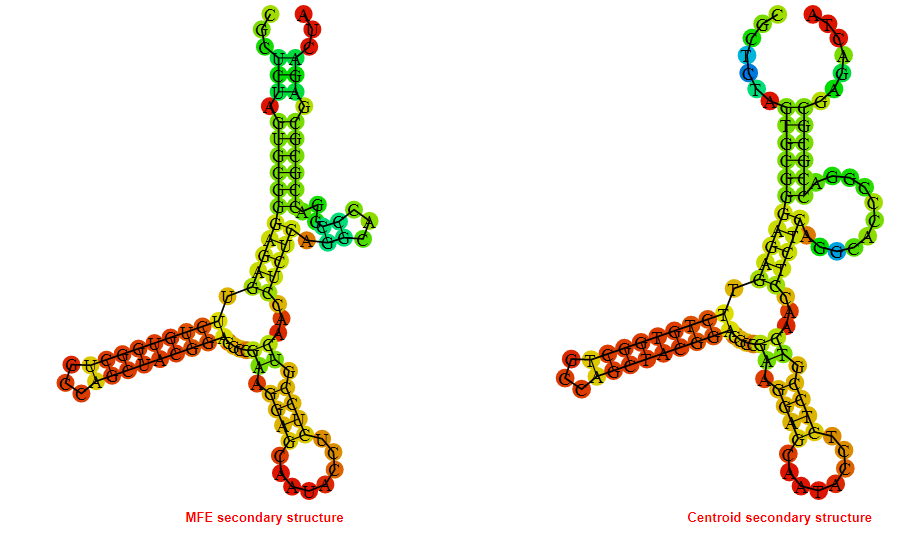
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Mycobacterium tuberculosis , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
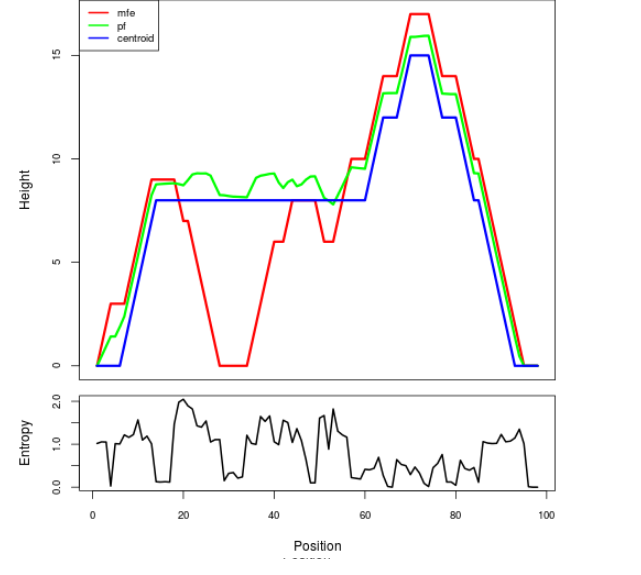
The plot represents the thermodynamic ensemble of the RNA structure of the Neisseria meningitidis , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
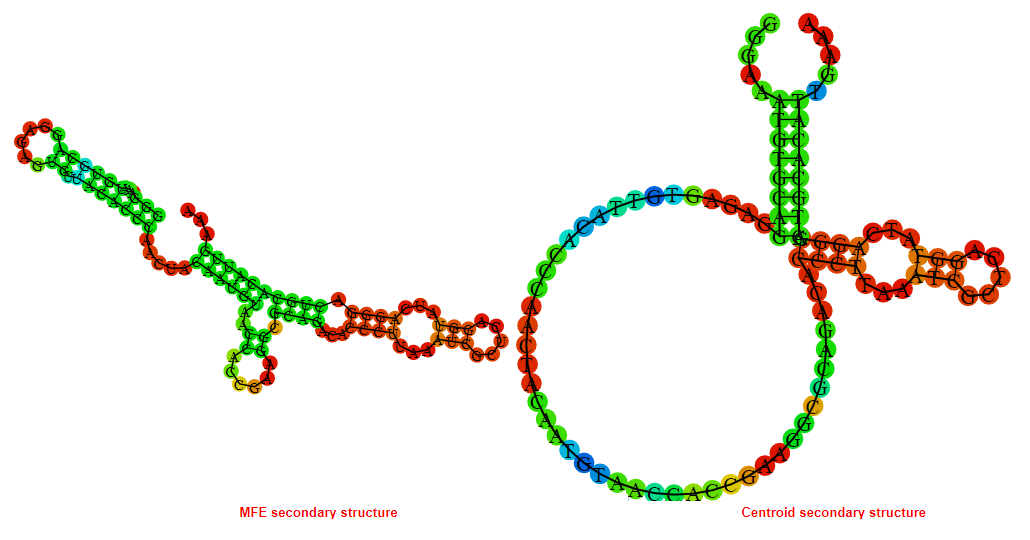
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Neisseria meningitidis , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
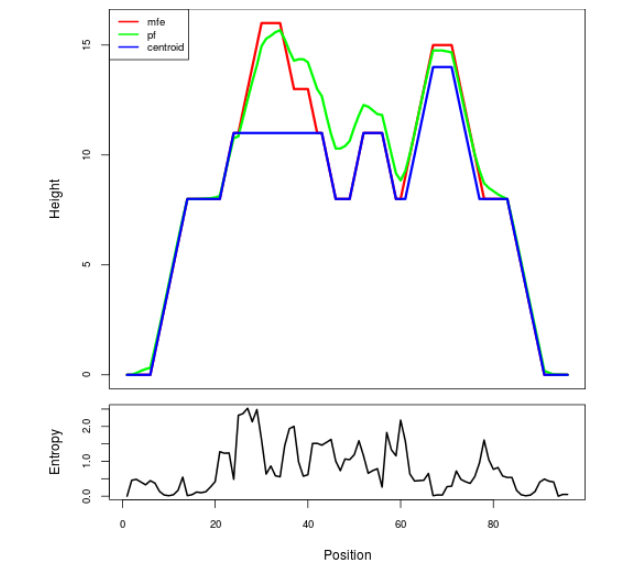
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus aureus , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
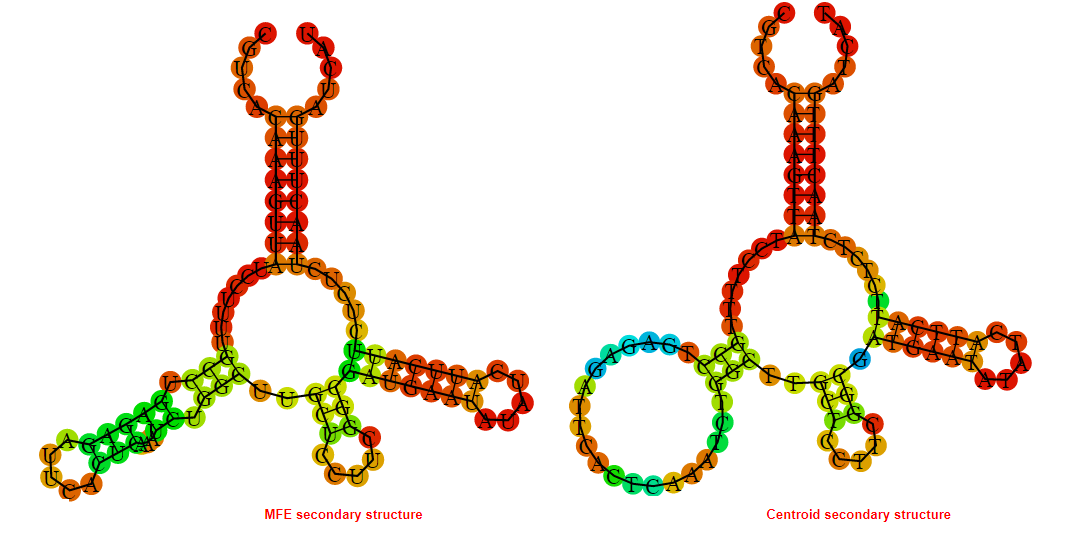
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus aureus , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
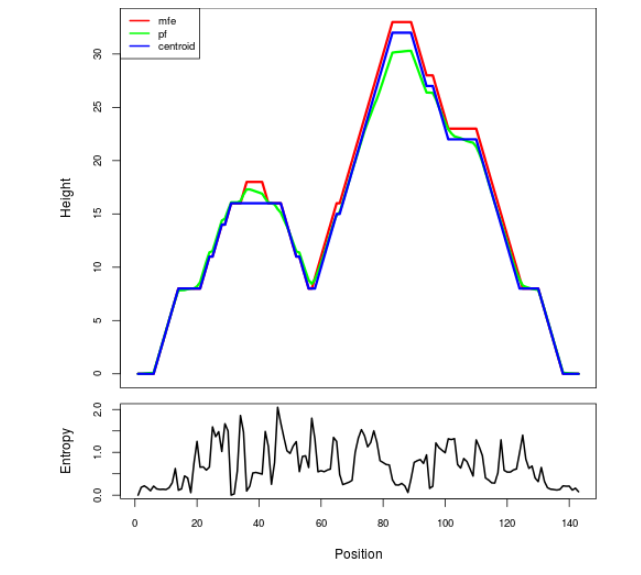
The plot represents the thermodynamic ensemble of the RNA structure of the Staphylococcus epidermidis , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
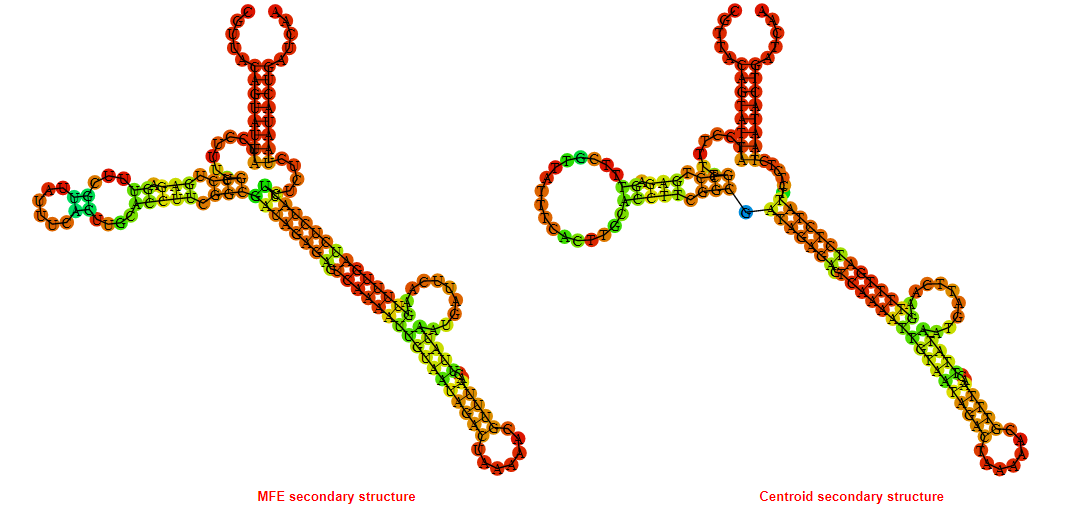
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Staphylococcus epidermidis , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
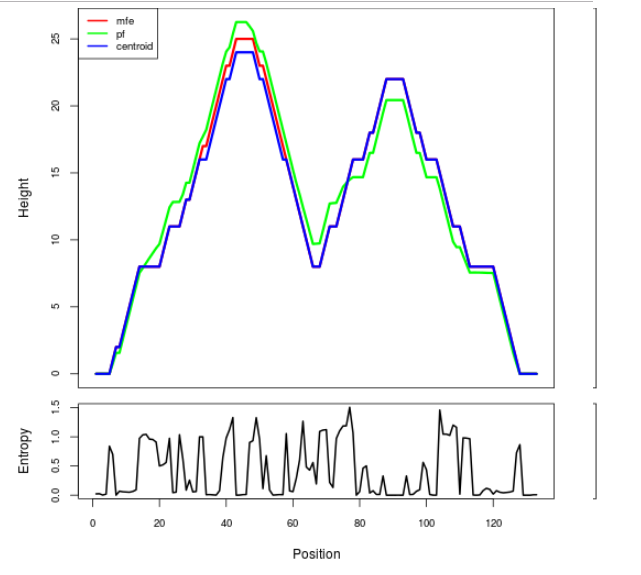
The plot represents the thermodynamic ensemble of the RNA structure of the Streptococcus agalactiae , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
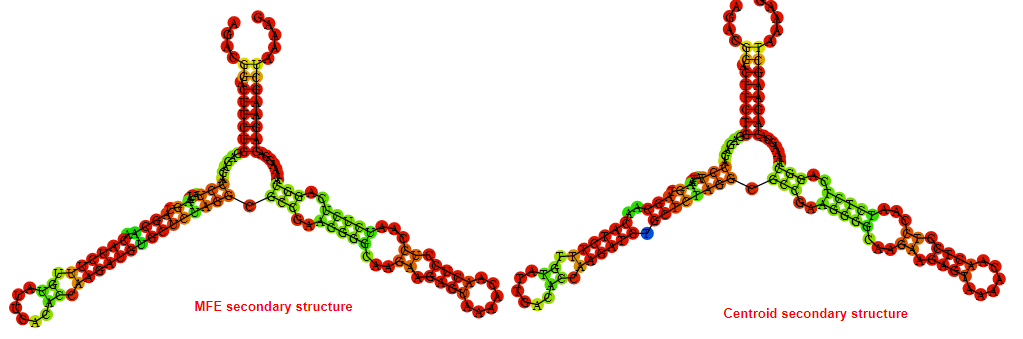
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Streptococcus agalactiae , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.

Multi_resistance strains for the Glycine type switches
The table displays known human pathogenic bacteria bearing the Glycine type switches. It includes the resistance of these
pathogens to currently available drugs. The first column provides the
name of the Human Bacterial Pathogen (HBP). The second one indicates
with a plus (+) symbol the presence of the riboswitch in the
particular bacterium. The third column indicates the resistance with a
(+) symbol to the Antiretroviral (AR) drug classes that are displayed
in the last column. In the HBP column, priority pathogens for the World Health Organisation are depicted in pink-filled box.
RiboSwitch_ID:
Back to the table
Go back to start a new search
RiboSwitch type
Bacteria strain target
Length of Switch in nucleotides
Sequence
Sequence information and analysis
Mountain plot representation of the switch structure
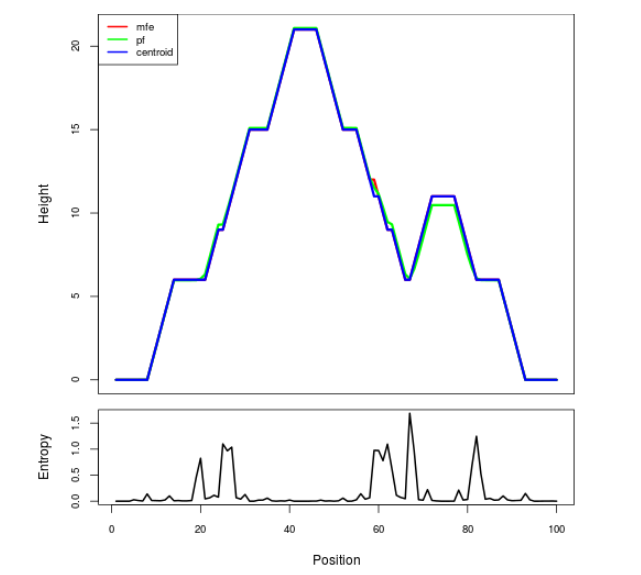
The plot represents the thermodynamic ensemble of the RNA structure of the Vibrio cholerae , Glycine type switch. In the plot the blue line indicates the predicted centroid structure of the switch. The red line represents the MFE predicted structure and the green line the thermodynamic ensemble of RNA structures. At the lower graph (the black curve), the positional entropy of each position on the switch sequence is displayed.
Free_Energy properties of the riboswitch
2D_Structure (A) (B)
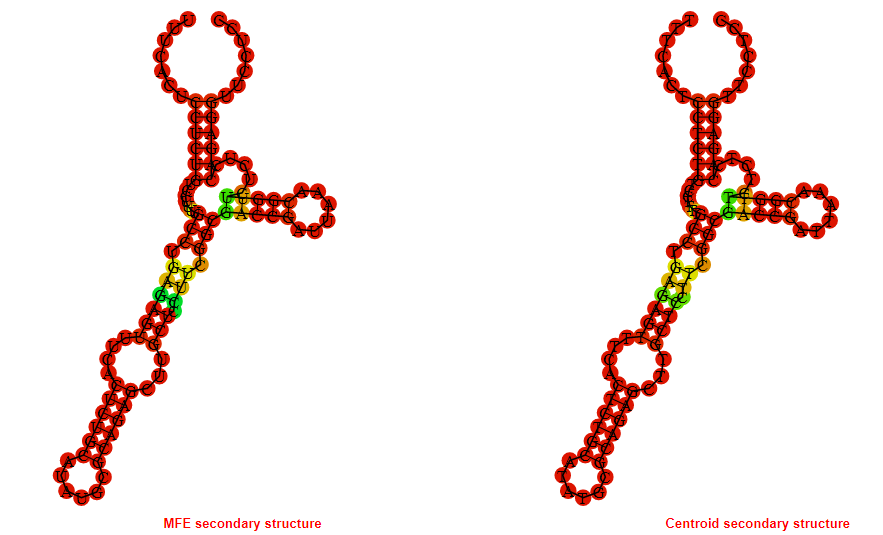
Display of the (A) MFE and (B) centroid secondary structures based on the thermodynamic properties presented above at the "Free_Energy" section of the Vibrio cholerae , Glycine type switch.
Multiple alignment of the Glycine type switches
Below is presented the sequences alignment of all the Glycine type switches currently present in the Rswitch database. In the alignment the consensus sequence is displayed and conserved regions amongst the sequences are highlighted. Less conserved nucleotides are also depicted and marked in squares. Coloring of the sequences generated with the ESPript3.x **
Motif_Search
The consensus sequence (general motif) of the Glycine type switch is presented below followed by motifs generated based on the consensus sequence. Moreover motifs extracted from the sequences alignment presented above in the "Multiple alignment" section are presented.
Key: N{0,4} = No, or up to 4 nucleotides can be present at the N position. N? = No or 1 nucleotide can be present at the N position.
The IUB ambiguity codes are : R = (G or A), Y = (C or T), M = (A or C), K = (G or T), S = (G or C), W = (A or T), B = (C or G or T), D = (A or G or T), H = (A or C or T), V = (A or C or G), N = (A or C or G or T)*
The Pathway which the Glycine type switches involved in.